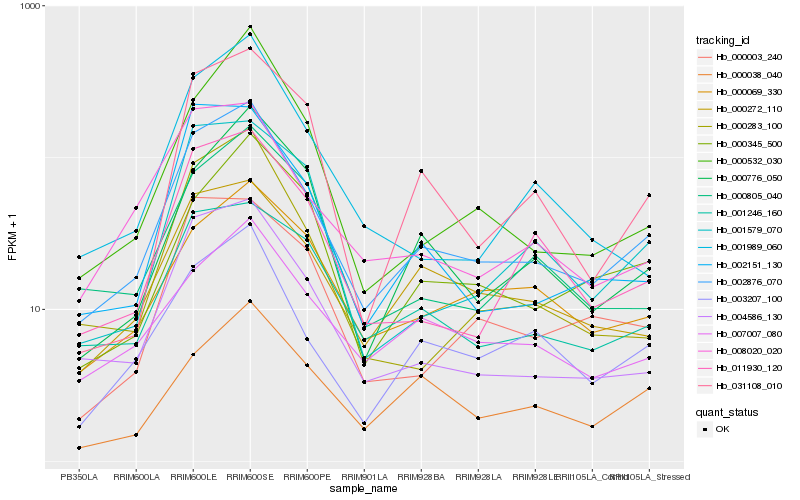
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_070 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 2 |
Hb_004586_130 |
0.1148872976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003207_100 |
0.1152283968 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 4 |
Hb_000272_110 |
0.1259989552 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011930_120 |
0.1300845208 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 6 |
Hb_008020_020 |
0.1329838828 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001246_160 |
0.1357872255 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 8 |
Hb_007007_080 |
0.1373790907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000776_050 |
0.1389316994 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 10 |
Hb_000345_500 |
0.1410940607 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 11 |
Hb_000283_100 |
0.1411840395 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000805_040 |
0.1414475023 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 13 |
Hb_000069_330 |
0.1424973257 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_002151_130 |
0.1428066643 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 15 |
Hb_000003_240 |
0.1472988332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001579_070 |
0.1476786119 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 17 |
Hb_001989_060 |
0.1490051401 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000038_040 |
0.1500923875 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 19 |
Hb_000532_030 |
0.1508461396 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 20 |
Hb_031108_010 |
0.150987279 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |