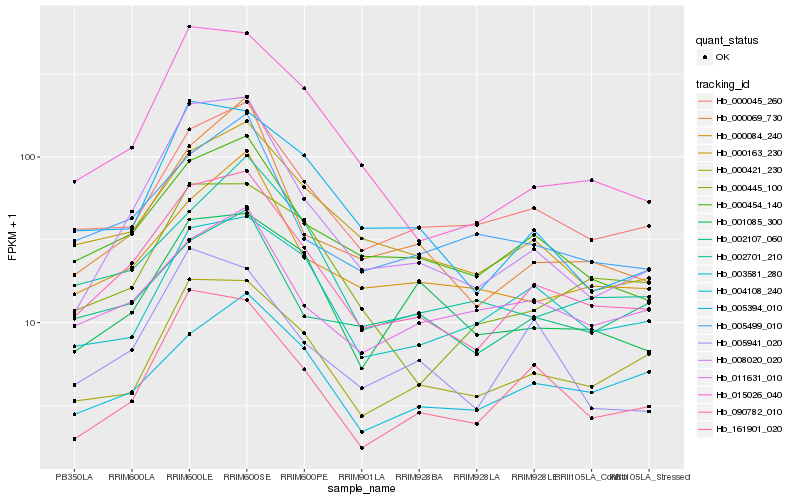
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090782_010 |
0.0 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 2 |
Hb_000454_140 |
0.09640034 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 3 |
Hb_000163_230 |
0.1020846646 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 4 |
Hb_000045_260 |
0.1057934899 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_008020_020 |
0.1159496635 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_161901_020 |
0.1189004776 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 7 |
Hb_015026_040 |
0.1189544188 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 8 |
Hb_000421_230 |
0.1194521756 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 9 |
Hb_005394_010 |
0.1246892401 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 10 |
Hb_000084_240 |
0.1267809416 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 11 |
Hb_000069_730 |
0.1285222268 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002701_210 |
0.1310268399 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 13 |
Hb_000445_100 |
0.1312540293 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 14 |
Hb_005499_010 |
0.1314839437 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 15 |
Hb_003581_280 |
0.1322271338 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 16 |
Hb_002107_060 |
0.1345154576 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 17 |
Hb_011631_010 |
0.1364985448 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 18 |
Hb_004108_240 |
0.1366043853 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 19 |
Hb_001085_300 |
0.1380917228 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 20 |
Hb_005941_020 |
0.1397376611 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |