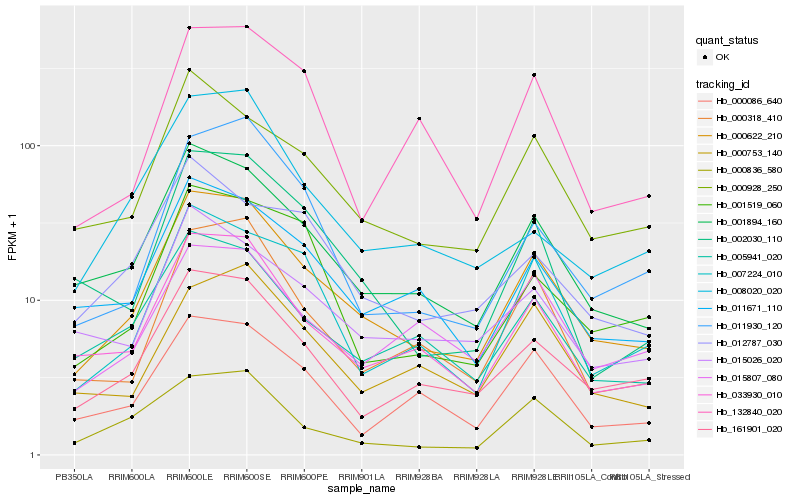
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_210 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_001894_160 |
0.0860360276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_033930_010 |
0.0980977497 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 4 |
Hb_161901_020 |
0.1055329092 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 5 |
Hb_000928_250 |
0.1139323499 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_000318_410 |
0.1151445845 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 7 |
Hb_005941_020 |
0.1164232858 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 8 |
Hb_011671_110 |
0.1177504473 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 9 |
Hb_007224_010 |
0.1237059731 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 10 |
Hb_002030_110 |
0.1304737129 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 11 |
Hb_000086_640 |
0.1311521671 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_000836_580 |
0.1333792088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_015026_020 |
0.1350605779 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 14 |
Hb_000753_140 |
0.1380806522 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 15 |
Hb_001519_060 |
0.1386696209 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_015807_080 |
0.1407038601 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 17 |
Hb_011930_120 |
0.1411039755 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 18 |
Hb_132840_020 |
0.1481072089 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 19 |
Hb_012787_030 |
0.1485620385 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 20 |
Hb_008020_020 |
0.1500735647 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |