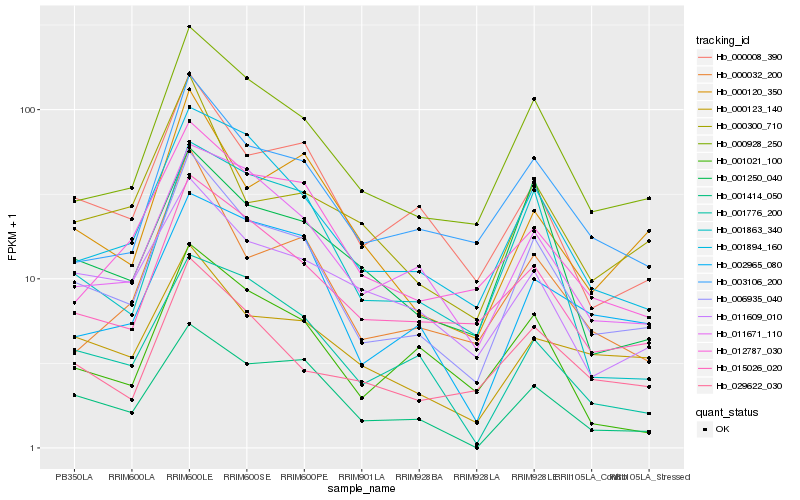
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006935_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 2 |
Hb_000928_250 |
0.0935337506 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_001894_160 |
0.1169438918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000008_390 |
0.1184039922 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 5 |
Hb_015026_020 |
0.1210590976 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 6 |
Hb_003106_200 |
0.1226018115 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001414_050 |
0.1244115147 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 8 |
Hb_011671_110 |
0.1298191096 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 9 |
Hb_000120_350 |
0.133507956 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 10 |
Hb_000123_140 |
0.1357288502 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 11 |
Hb_000032_200 |
0.1357558307 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 12 |
Hb_012787_030 |
0.1400527032 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 13 |
Hb_029622_030 |
0.1414409038 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001250_040 |
0.144634396 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 15 |
Hb_002965_080 |
0.1453634258 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 16 |
Hb_000300_710 |
0.1462689421 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 17 |
Hb_011609_010 |
0.1466111022 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 18 |
Hb_001776_200 |
0.1499332948 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 19 |
Hb_001863_340 |
0.1510411499 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 20 |
Hb_001021_100 |
0.1520541014 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |