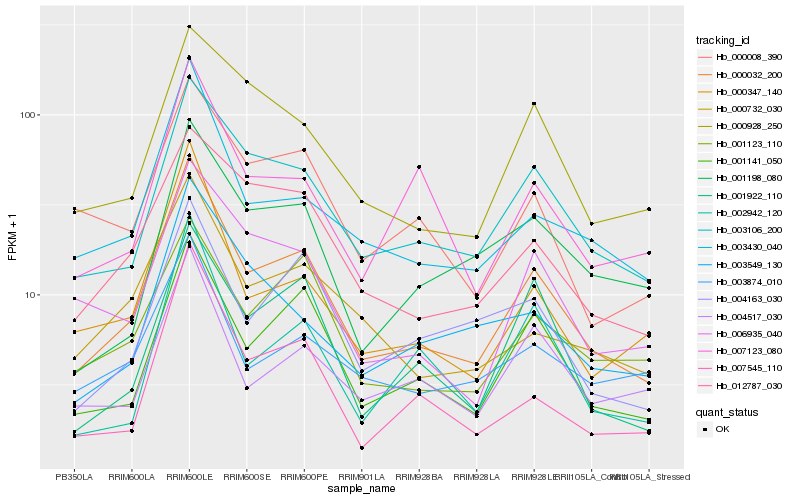
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_200 |
0.0 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 2 |
Hb_001141_050 |
0.1044208842 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003106_200 |
0.1082188122 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003430_040 |
0.1192850621 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 5 |
Hb_000347_140 |
0.1244558683 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_012787_030 |
0.1303676422 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 7 |
Hb_000732_030 |
0.1312748339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007545_110 |
0.1333531861 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_006935_040 |
0.1357558307 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 10 |
Hb_001922_110 |
0.1406131444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004163_030 |
0.1425711241 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 12 |
Hb_001123_110 |
0.1426309682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000008_390 |
0.1433883592 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 14 |
Hb_000928_250 |
0.143528486 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_004517_030 |
0.1447229528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002942_120 |
0.1469230084 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 17 |
Hb_007123_080 |
0.1477167676 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 18 |
Hb_001198_080 |
0.1484504497 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003549_130 |
0.1498158997 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 20 |
Hb_003874_010 |
0.1500813864 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |