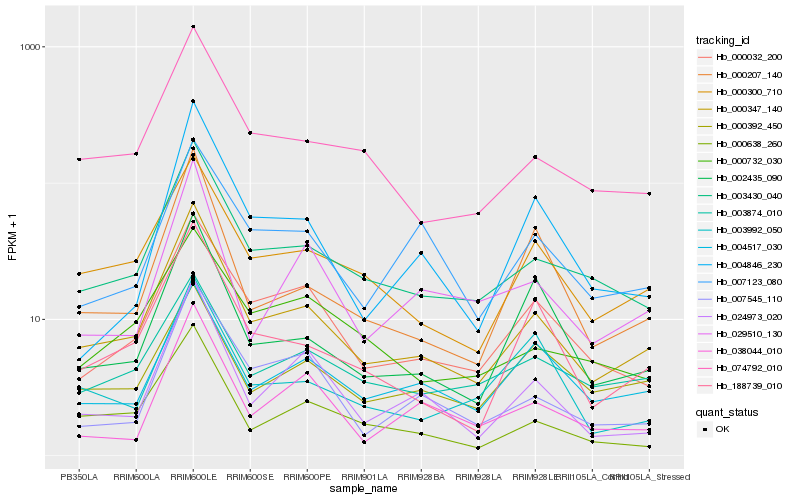
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_140 |
0.0 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_003430_040 |
0.0964183502 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 3 |
Hb_074792_010 |
0.1099009176 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 4 |
Hb_000300_710 |
0.1099622948 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 5 |
Hb_188739_010 |
0.1236793948 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000032_200 |
0.1244558683 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_000638_260 |
0.1316639407 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 8 |
Hb_003874_010 |
0.1370285517 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000207_140 |
0.1389313847 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 10 |
Hb_024973_020 |
0.1396430792 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 11 |
Hb_004517_030 |
0.1402047243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002435_090 |
0.1407444366 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 13 |
Hb_007545_110 |
0.1454624003 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_000392_450 |
0.1472303245 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_029510_130 |
0.1520849177 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 16 |
Hb_000732_030 |
0.153350172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007123_080 |
0.1540356093 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 18 |
Hb_004846_230 |
0.1608657434 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_003992_050 |
0.1611340858 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 20 |
Hb_038044_010 |
0.1619069092 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |