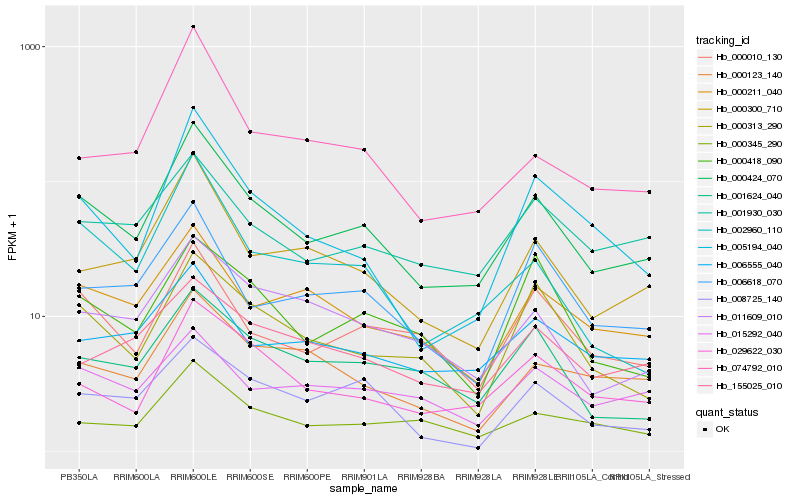
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_070 |
0.0 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 2 |
Hb_000300_710 |
0.1274557534 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 3 |
Hb_029622_030 |
0.1276365533 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000010_130 |
0.1303663599 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic [Pyrus x bretschneideri] |
| 5 |
Hb_000211_040 |
0.1358769689 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 6 |
Hb_002960_110 |
0.1368401134 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005194_040 |
0.1369394631 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_001930_030 |
0.1395262218 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 9 |
Hb_011609_010 |
0.1409839819 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 10 |
Hb_006618_070 |
0.141502395 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000345_290 |
0.141888157 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000313_290 |
0.1438374545 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_006555_040 |
0.1439459871 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 14 |
Hb_001624_040 |
0.1457438311 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000123_140 |
0.1462942332 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 16 |
Hb_008725_140 |
0.1514191698 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_074792_010 |
0.1519931313 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 18 |
Hb_015292_040 |
0.1543574136 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 19 |
Hb_155025_010 |
0.1573827024 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 20 |
Hb_000418_090 |
0.1578373627 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |