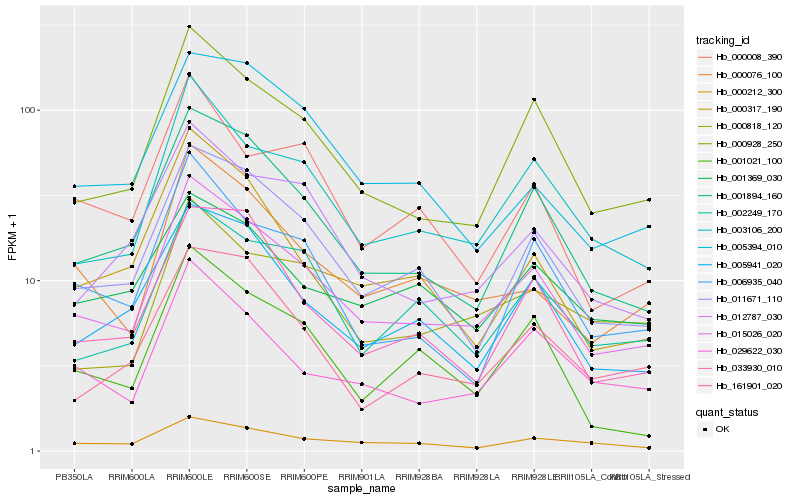
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015026_020 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 2 |
Hb_000928_250 |
0.0803872661 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_001894_160 |
0.0837881559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011671_110 |
0.0887350527 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 5 |
Hb_003106_200 |
0.0892673878 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005941_020 |
0.1040057004 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 7 |
Hb_000076_100 |
0.1060321468 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_012787_030 |
0.1120583708 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 9 |
Hb_029622_030 |
0.1132015508 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000212_300 |
0.1147695636 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 11 |
Hb_002249_170 |
0.11626957 |
- |
- |
copine, putative [Ricinus communis] |
| 12 |
Hb_000317_190 |
0.1190471109 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 13 |
Hb_033930_010 |
0.1205462082 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 14 |
Hb_006935_040 |
0.1210590976 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 15 |
Hb_001021_100 |
0.1253114125 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 16 |
Hb_000818_120 |
0.1259891111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001369_030 |
0.1281554562 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 18 |
Hb_161901_020 |
0.1304306529 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 19 |
Hb_000008_390 |
0.1320974573 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 20 |
Hb_005394_010 |
0.1324026406 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |