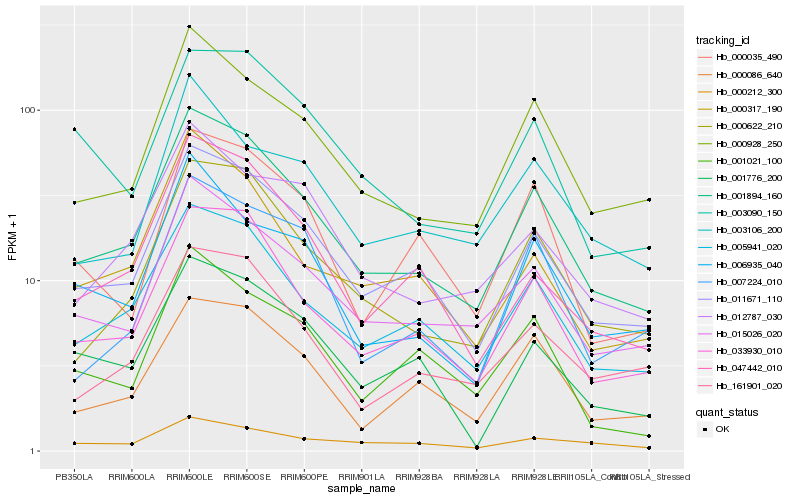
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001894_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011671_110 |
0.0638661315 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 3 |
Hb_005941_020 |
0.069322137 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 4 |
Hb_033930_010 |
0.0748741022 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 5 |
Hb_000928_250 |
0.0805019655 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_015026_020 |
0.0837881559 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 7 |
Hb_000622_210 |
0.0860360276 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_161901_020 |
0.1089527433 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 9 |
Hb_000086_640 |
0.1097650532 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_012787_030 |
0.1150410974 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 11 |
Hb_000317_190 |
0.1151529968 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 12 |
Hb_047442_010 |
0.1168267584 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 13 |
Hb_006935_040 |
0.1169438918 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 14 |
Hb_000212_300 |
0.1211053911 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 15 |
Hb_003106_200 |
0.1240189515 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003090_150 |
0.1278676933 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 17 |
Hb_001776_200 |
0.1278779469 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 18 |
Hb_007224_010 |
0.1281210669 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 19 |
Hb_000035_490 |
0.1284075994 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 20 |
Hb_001021_100 |
0.1284963851 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |