| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
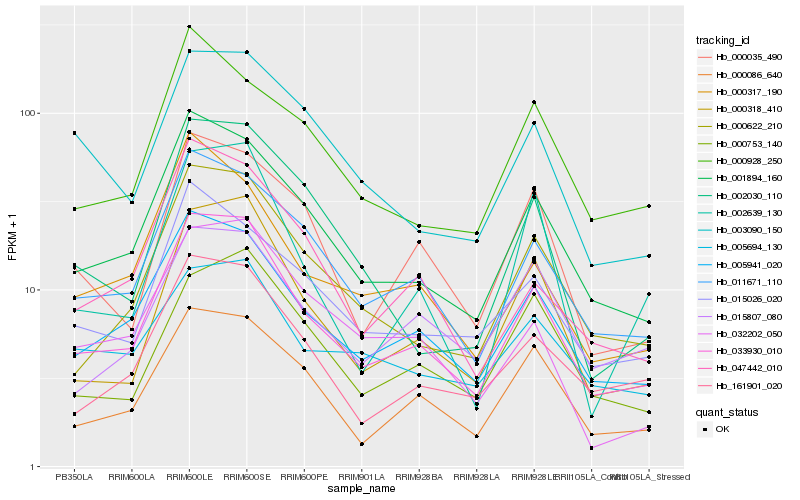
Hb_033930_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 2 |
Hb_005941_020 |
0.0727475297 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 3 |
Hb_001894_160 |
0.0748741022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011671_110 |
0.0853342528 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 5 |
Hb_000318_410 |
0.0947957082 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 6 |
Hb_000622_210 |
0.0980977497 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 7 |
Hb_000086_640 |
0.1024243373 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_161901_020 |
0.1069361575 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 9 |
Hb_015026_020 |
0.1205462082 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 10 |
Hb_047442_010 |
0.121386815 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 11 |
Hb_000035_490 |
0.1214623375 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 12 |
Hb_000753_140 |
0.1261729149 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 13 |
Hb_000317_190 |
0.1280114045 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 14 |
Hb_032202_050 |
0.1286726553 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005694_130 |
0.1317613292 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 16 |
Hb_015807_080 |
0.1330332077 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 17 |
Hb_003090_150 |
0.1338601603 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 18 |
Hb_000928_250 |
0.1348995997 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_002030_110 |
0.1351845549 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 20 |
Hb_002639_130 |
0.1366663392 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |