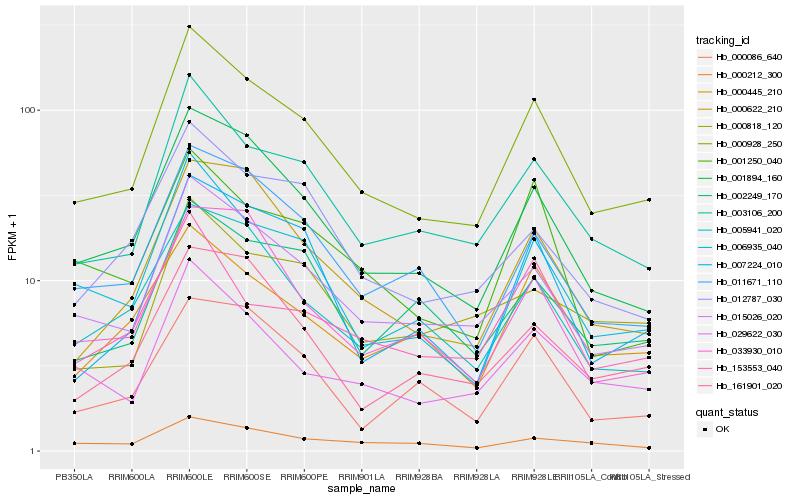
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_250 |
0.0 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_003106_200 |
0.0803703474 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 3 |
Hb_015026_020 |
0.0803872661 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 4 |
Hb_001894_160 |
0.0805019655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006935_040 |
0.0935337506 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 6 |
Hb_011671_110 |
0.1099422785 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 7 |
Hb_000622_210 |
0.1139323499 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_029622_030 |
0.1141206715 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_012787_030 |
0.1150875355 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 10 |
Hb_007224_010 |
0.1176899896 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 11 |
Hb_005941_020 |
0.1199618463 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 12 |
Hb_000445_210 |
0.123384801 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 13 |
Hb_002249_170 |
0.1269696694 |
- |
- |
copine, putative [Ricinus communis] |
| 14 |
Hb_001250_040 |
0.1312159075 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 15 |
Hb_161901_020 |
0.1314411258 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 16 |
Hb_153553_040 |
0.1336887351 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 17 |
Hb_033930_010 |
0.1348995997 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 18 |
Hb_000212_300 |
0.1358288492 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 19 |
Hb_000086_640 |
0.1376542383 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_000818_120 |
0.1384431412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |