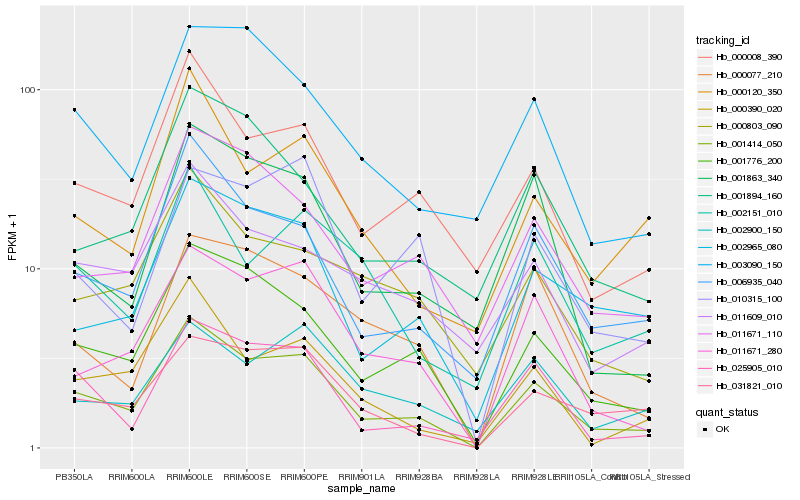
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001414_050 |
0.0 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 2 |
Hb_001776_200 |
0.1092289301 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 3 |
Hb_006935_040 |
0.1244115147 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 4 |
Hb_000008_390 |
0.1331064092 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 5 |
Hb_011671_280 |
0.1345103889 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF106-like [Populus euphratica] |
| 6 |
Hb_031821_010 |
0.1402458496 |
- |
- |
ccd1, putative [Ricinus communis] |
| 7 |
Hb_002965_080 |
0.1447305393 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 8 |
Hb_025905_010 |
0.1449256926 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 9 |
Hb_011671_110 |
0.1504880086 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 10 |
Hb_000390_020 |
0.1545374207 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 11 |
Hb_001863_340 |
0.1586378845 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 12 |
Hb_000803_090 |
0.1597495909 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 13 |
Hb_011609_010 |
0.1605615832 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 14 |
Hb_000120_350 |
0.1626672727 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 15 |
Hb_000077_210 |
0.1627883374 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 16 |
Hb_010315_100 |
0.1656938324 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 17 |
Hb_003090_150 |
0.1665328436 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 18 |
Hb_002151_010 |
0.1675702597 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 19 |
Hb_001894_160 |
0.1675810898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002900_150 |
0.1688300506 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |