| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
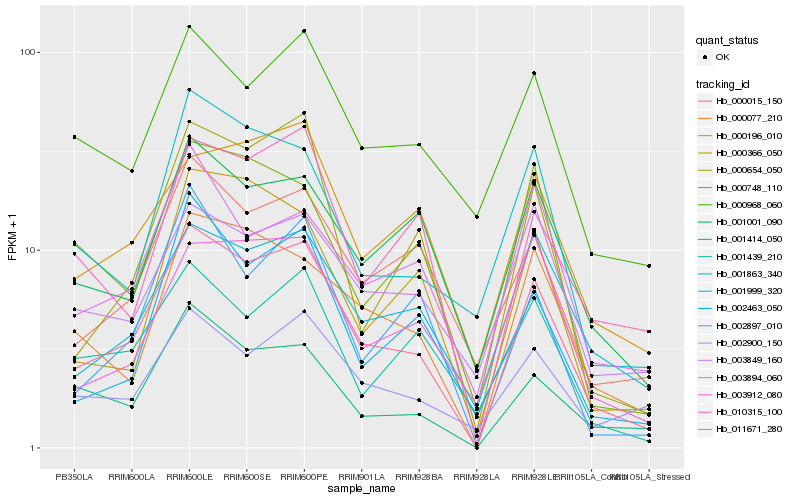
Hb_011671_280 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF106-like [Populus euphratica] |
| 2 |
Hb_001414_050 |
0.1345103889 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 3 |
Hb_000077_210 |
0.1360128702 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 4 |
Hb_000015_150 |
0.1415882097 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 5 |
Hb_000654_050 |
0.1442630574 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 6 |
Hb_001999_320 |
0.1460014546 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 7 |
Hb_000196_010 |
0.1499201551 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 8 |
Hb_001001_090 |
0.1499991013 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 9 |
Hb_000968_060 |
0.1521199327 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 10 |
Hb_002463_050 |
0.1524122187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010315_100 |
0.1534075899 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 12 |
Hb_001863_340 |
0.1545184105 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 13 |
Hb_002900_150 |
0.1546357116 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 14 |
Hb_003912_080 |
0.158988538 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 15 |
Hb_000366_050 |
0.1593304658 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_002897_010 |
0.1598332734 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 17 |
Hb_001439_210 |
0.1607005332 |
- |
- |
EXS family protein [Populus trichocarpa] |
| 18 |
Hb_000748_110 |
0.1610556456 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 19 |
Hb_003849_160 |
0.1615366757 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 20 |
Hb_003894_060 |
0.1634084484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |