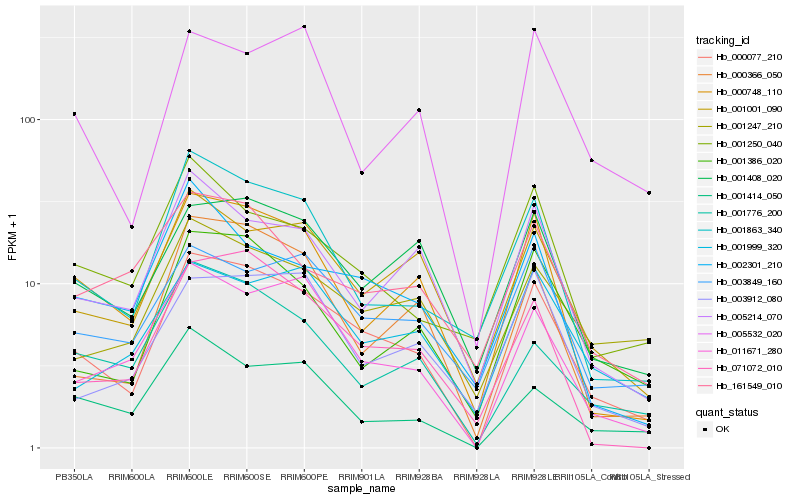
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_210 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 2 |
Hb_000748_110 |
0.1347501216 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 3 |
Hb_011671_280 |
0.1360128702 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF106-like [Populus euphratica] |
| 4 |
Hb_001863_340 |
0.1439707467 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 5 |
Hb_001001_090 |
0.1443148415 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 6 |
Hb_000366_050 |
0.1447488052 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_001386_020 |
0.1457315768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001408_020 |
0.1491989925 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 9 |
Hb_005214_070 |
0.1530434552 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 10 |
Hb_003849_160 |
0.1559647717 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 11 |
Hb_071072_010 |
0.1561042131 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_001776_200 |
0.1614888188 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 13 |
Hb_001247_210 |
0.1618731275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001414_050 |
0.1627883374 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 15 |
Hb_001999_320 |
0.1648795764 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 16 |
Hb_002301_210 |
0.1661952006 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003912_080 |
0.1667048051 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 18 |
Hb_005532_020 |
0.167688959 |
- |
- |
PREDICTED: uncharacterized protein LOC100262861 [Vitis vinifera] |
| 19 |
Hb_001250_040 |
0.1687136384 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 20 |
Hb_161549_010 |
0.1687214031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |