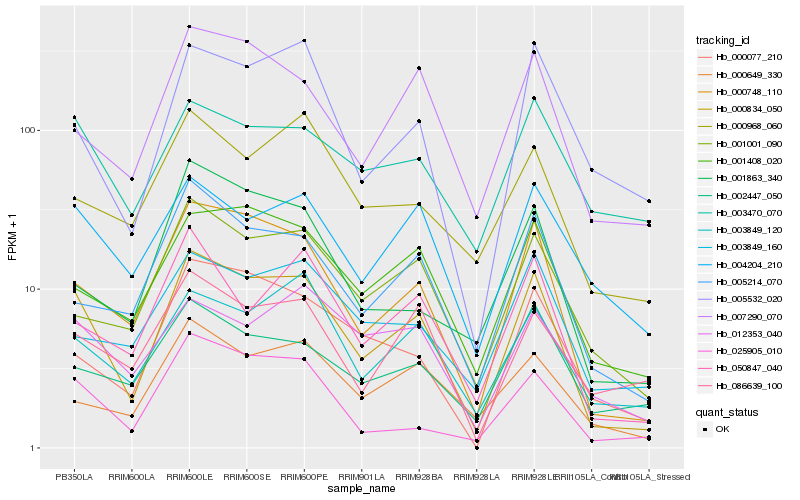
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_050 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 2 |
Hb_000748_110 |
0.1129902585 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 3 |
Hb_025905_010 |
0.1555690564 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_001408_020 |
0.1559284407 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 5 |
Hb_005214_070 |
0.1573598025 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 6 |
Hb_003849_120 |
0.1610736899 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 7 |
Hb_007290_070 |
0.1639321102 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 8 |
Hb_001001_090 |
0.1642711128 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 9 |
Hb_005532_020 |
0.1646075713 |
- |
- |
PREDICTED: uncharacterized protein LOC100262861 [Vitis vinifera] |
| 10 |
Hb_050847_040 |
0.166439282 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 11 |
Hb_004204_210 |
0.1686973137 |
- |
- |
PREDICTED: heparanase-like protein 1 [Jatropha curcas] |
| 12 |
Hb_000077_210 |
0.169929681 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 13 |
Hb_012353_040 |
0.1740469948 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_000649_330 |
0.1743906312 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 15 |
Hb_086639_100 |
0.1762237132 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105646787 [Jatropha curcas] |
| 16 |
Hb_002447_050 |
0.1803980041 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_003849_160 |
0.1812527978 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 18 |
Hb_003470_070 |
0.1822805334 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 19 |
Hb_000968_060 |
0.1824807565 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 20 |
Hb_001863_340 |
0.1848687401 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |