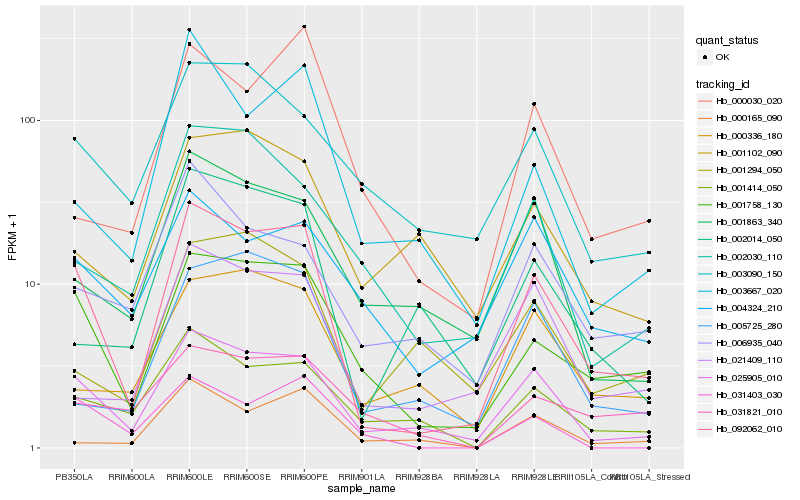
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092062_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 2 |
Hb_025905_010 |
0.1356803477 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 3 |
Hb_001758_130 |
0.1463108006 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 4 |
Hb_001414_050 |
0.2081450399 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 5 |
Hb_031403_030 |
0.2193844089 |
- |
- |
- |
| 6 |
Hb_021409_110 |
0.2225558254 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_003667_020 |
0.2227451952 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 8 |
Hb_001863_340 |
0.2246385919 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 9 |
Hb_003090_150 |
0.2267166314 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 10 |
Hb_002030_110 |
0.2291995568 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 11 |
Hb_002014_050 |
0.229471978 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 12 |
Hb_006935_040 |
0.2296193561 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 13 |
Hb_005725_280 |
0.2316818021 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 14 |
Hb_000336_180 |
0.2317305919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_031821_010 |
0.2349759945 |
- |
- |
ccd1, putative [Ricinus communis] |
| 16 |
Hb_004324_210 |
0.2352260776 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 17 |
Hb_000030_020 |
0.2362132792 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 18 |
Hb_000165_090 |
0.2390611592 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 2-like [Jatropha curcas] |
| 19 |
Hb_001102_090 |
0.2419047084 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 20 |
Hb_001294_050 |
0.2427569041 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |