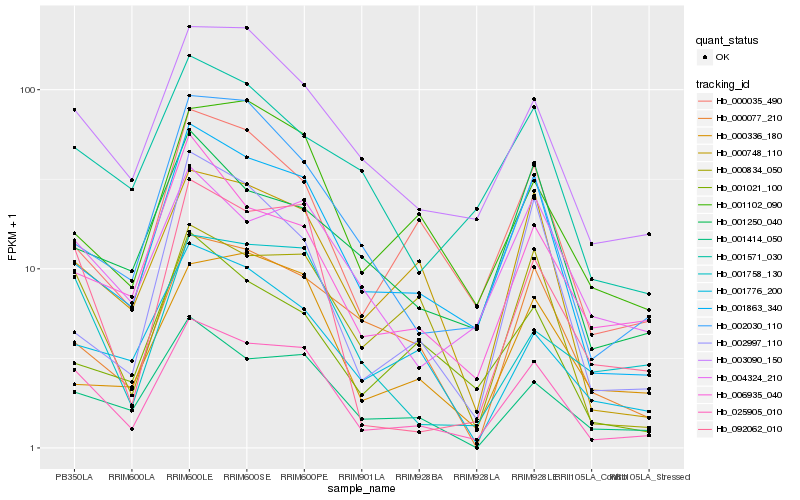
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025905_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 2 |
Hb_001863_340 |
0.1295145494 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 3 |
Hb_092062_010 |
0.1356803477 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 4 |
Hb_001414_050 |
0.1449256926 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 5 |
Hb_003090_150 |
0.1474148843 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 6 |
Hb_000834_050 |
0.1555690564 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 7 |
Hb_000748_110 |
0.156240739 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 8 |
Hb_000035_490 |
0.1699171786 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 9 |
Hb_001758_130 |
0.1703636846 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 10 |
Hb_004324_210 |
0.1730757134 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 11 |
Hb_002030_110 |
0.1739595616 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 12 |
Hb_006935_040 |
0.1749271618 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 13 |
Hb_001102_090 |
0.1793393409 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 14 |
Hb_001776_200 |
0.1829237298 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 15 |
Hb_001571_030 |
0.1858823432 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001250_040 |
0.1860946255 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 17 |
Hb_000336_180 |
0.1862086558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000077_210 |
0.1893640998 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 19 |
Hb_002997_110 |
0.1901246166 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001021_100 |
0.1907153527 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |