| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
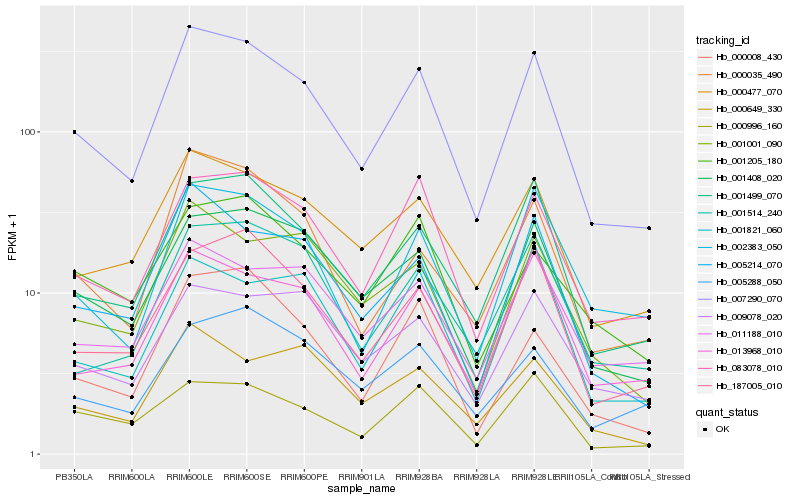
Hb_007290_070 |
0.0 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 2 |
Hb_001499_070 |
0.0876637448 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 3 |
Hb_001408_020 |
0.0925732213 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 4 |
Hb_083078_010 |
0.0929296323 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 5 |
Hb_000477_070 |
0.0969850351 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 6 |
Hb_005214_070 |
0.1010747373 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 7 |
Hb_187005_010 |
0.101820143 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 8 |
Hb_002383_050 |
0.1061464635 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 9 |
Hb_013968_010 |
0.1097657093 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 10 |
Hb_011188_010 |
0.1122506683 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001001_090 |
0.1139169481 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 12 |
Hb_000035_490 |
0.1160719239 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 13 |
Hb_001514_240 |
0.1176586175 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 14 |
Hb_000996_160 |
0.1181271188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009078_020 |
0.1189594037 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 16 |
Hb_000008_430 |
0.1223573359 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005288_050 |
0.1269978181 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 18 |
Hb_000649_330 |
0.1270780835 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 19 |
Hb_001205_180 |
0.1272795692 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 20 |
Hb_001821_060 |
0.1281195899 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |