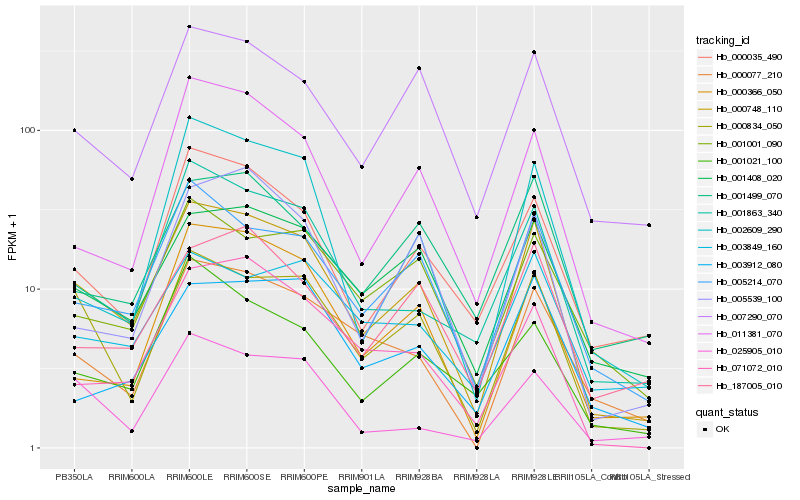
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000748_110 |
0.0 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 2 |
Hb_005214_070 |
0.1104168851 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 3 |
Hb_000834_050 |
0.1129902585 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 4 |
Hb_001408_020 |
0.1225849868 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 5 |
Hb_001863_340 |
0.1269349074 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 6 |
Hb_007290_070 |
0.1289023167 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 7 |
Hb_000077_210 |
0.1347501216 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 8 |
Hb_000035_490 |
0.1366002255 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 9 |
Hb_011381_070 |
0.1383465641 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 10 |
Hb_001001_090 |
0.1395392514 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 11 |
Hb_000366_050 |
0.1437628918 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 12 |
Hb_187005_010 |
0.1465982367 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 13 |
Hb_071072_010 |
0.1496382218 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_005539_100 |
0.1516230136 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 15 |
Hb_001499_070 |
0.1529102944 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 16 |
Hb_003849_160 |
0.1549503452 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 17 |
Hb_002609_290 |
0.1553211017 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 18 |
Hb_001021_100 |
0.1557468454 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 19 |
Hb_003912_080 |
0.1560504466 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 20 |
Hb_025905_010 |
0.156240739 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |