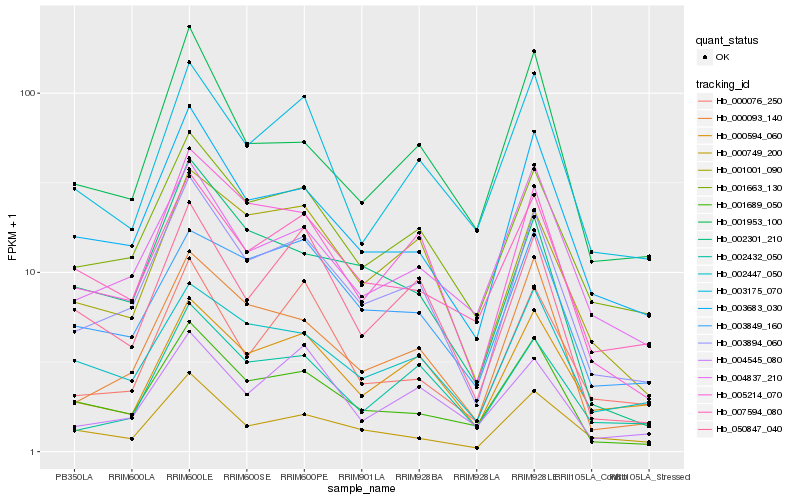
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003894_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001663_130 |
0.0828692518 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 3 |
Hb_003683_030 |
0.0903077053 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 4 |
Hb_005214_070 |
0.0916531334 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 5 |
Hb_000093_140 |
0.1057667419 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001001_090 |
0.1086535626 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 7 |
Hb_002432_050 |
0.1136837134 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 8 |
Hb_001689_050 |
0.1141267874 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 9 |
Hb_000594_060 |
0.1144286666 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 10 |
Hb_002447_050 |
0.1184585492 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_004837_210 |
0.1220592528 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 12 |
Hb_007594_080 |
0.1229372063 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 13 |
Hb_050847_040 |
0.125602197 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 14 |
Hb_003849_160 |
0.1261669254 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 15 |
Hb_003175_070 |
0.1265008768 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 16 |
Hb_000749_200 |
0.1274852651 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004545_080 |
0.1284093783 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_001953_100 |
0.1293619887 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002301_210 |
0.129822253 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000076_250 |
0.1298439186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |