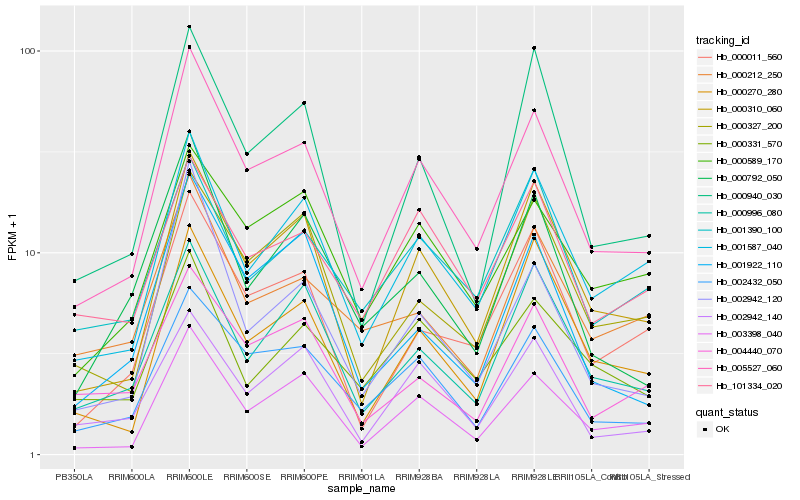
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005527_060 |
0.0 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 2 |
Hb_002432_050 |
0.0877523598 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 3 |
Hb_000310_060 |
0.0919668624 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 4 |
Hb_003398_040 |
0.0979635641 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001587_040 |
0.0998127227 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000589_170 |
0.1148889636 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000011_560 |
0.1177530413 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000996_080 |
0.1206001674 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 9 |
Hb_000940_030 |
0.1220058865 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000327_200 |
0.1242193077 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 11 |
Hb_002942_140 |
0.1245592002 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 12 |
Hb_101334_020 |
0.1256801826 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 13 |
Hb_002942_120 |
0.1272122035 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 14 |
Hb_000792_050 |
0.128382684 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000331_570 |
0.1296063482 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 16 |
Hb_000212_250 |
0.1300843013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000270_280 |
0.1303324166 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 18 |
Hb_004440_070 |
0.1307578015 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520 [Jatropha curcas] |
| 19 |
Hb_001390_100 |
0.1315159682 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001922_110 |
0.1316467977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |