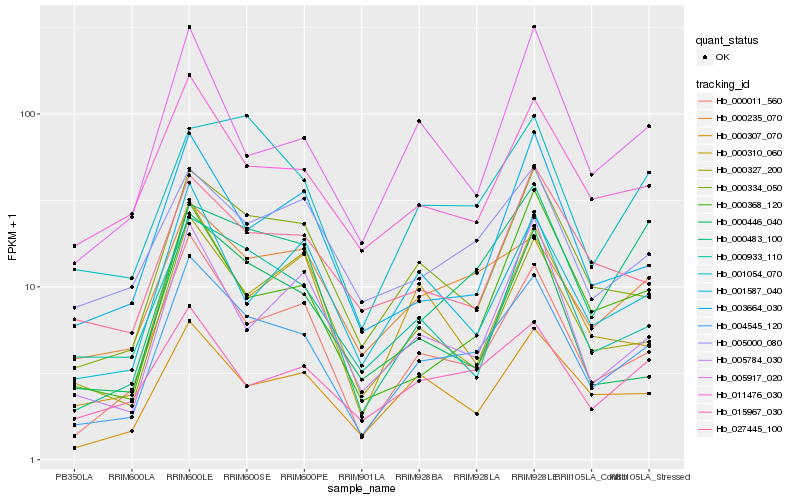
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004545_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647604 [Jatropha curcas] |
| 2 |
Hb_000011_560 |
0.097382702 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000334_050 |
0.1210779757 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 4 |
Hb_000235_070 |
0.1281140662 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 5 |
Hb_000307_070 |
0.1330746471 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_011476_030 |
0.1361262476 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 7 |
Hb_015967_030 |
0.1481961525 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000483_100 |
0.1484622542 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 9 |
Hb_000327_200 |
0.1491627912 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 10 |
Hb_000446_040 |
0.1508302086 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 11 |
Hb_005000_080 |
0.1513790654 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 12 |
Hb_000933_110 |
0.1514748786 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 13 |
Hb_001587_040 |
0.151877018 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001054_070 |
0.1535933025 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 15 |
Hb_005917_020 |
0.1536088242 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 16 |
Hb_003664_030 |
0.1542845137 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 17 |
Hb_027445_100 |
0.1561730313 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 18 |
Hb_005784_030 |
0.1579865228 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000310_060 |
0.1580459635 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 20 |
Hb_000368_120 |
0.1584159142 |
- |
- |
PREDICTED: uncharacterized protein LOC105633862 [Jatropha curcas] |