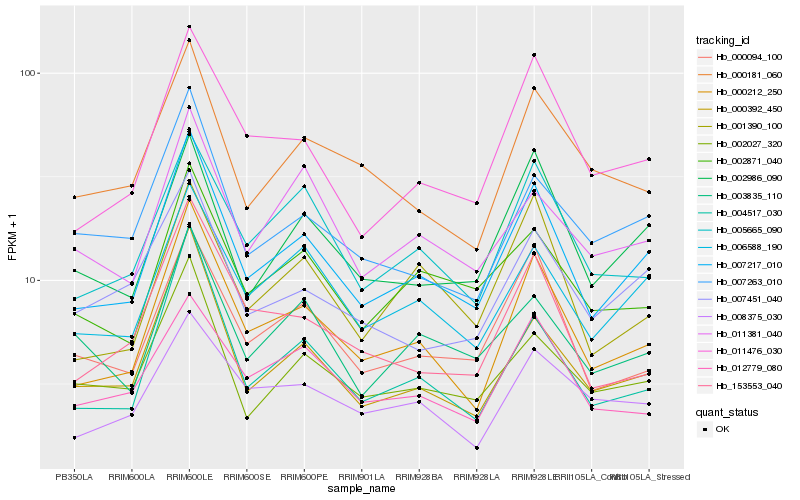
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007217_010 |
0.0 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000212_250 |
0.0693062625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006588_190 |
0.0865753478 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 4 |
Hb_011476_030 |
0.0930347344 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 5 |
Hb_002986_090 |
0.0940050154 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_000392_450 |
0.0961886492 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000094_100 |
0.0966686308 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 8 |
Hb_002027_320 |
0.1011273047 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 9 |
Hb_005665_090 |
0.1029093133 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 10 |
Hb_002871_040 |
0.1034798473 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 11 |
Hb_004517_030 |
0.1045802418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007451_040 |
0.1087593137 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 13 |
Hb_007263_010 |
0.1095779109 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 14 |
Hb_001390_100 |
0.1096998298 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_153553_040 |
0.1150730255 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 16 |
Hb_003835_110 |
0.1156016721 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011381_040 |
0.1163234653 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000181_060 |
0.1170223936 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 19 |
Hb_012779_080 |
0.1175921918 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 20 |
Hb_008375_030 |
0.1176941179 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |