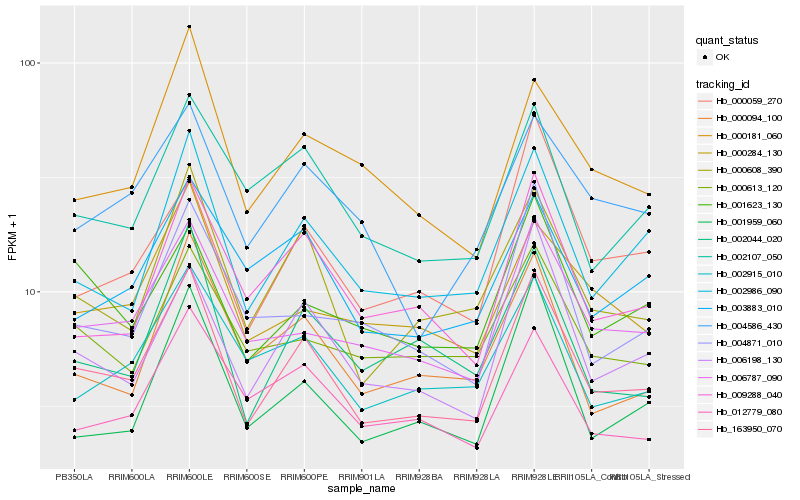
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163950_070 |
0.0 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 2 |
Hb_000608_390 |
0.088699685 |
- |
- |
hypothetical protein B456_010G140700 [Gossypium raimondii] |
| 3 |
Hb_002986_090 |
0.096767554 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 4 |
Hb_006198_130 |
0.0982776971 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 5 |
Hb_006787_090 |
0.1014443139 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 6 |
Hb_002044_020 |
0.1119111456 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 7 |
Hb_009288_040 |
0.1119697483 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 8 |
Hb_000094_100 |
0.1125082836 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 9 |
Hb_002107_050 |
0.1171355003 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 10 |
Hb_002915_010 |
0.1173335446 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 11 |
Hb_001959_060 |
0.1181714302 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000181_060 |
0.1196891269 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004586_430 |
0.1199171932 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001623_130 |
0.120285521 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000284_130 |
0.1207056072 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 16 |
Hb_012779_080 |
0.1207698929 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000613_120 |
0.1225163048 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004871_010 |
0.1229988775 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 19 |
Hb_003883_010 |
0.1240137789 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 20 |
Hb_000059_270 |
0.1268020057 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |