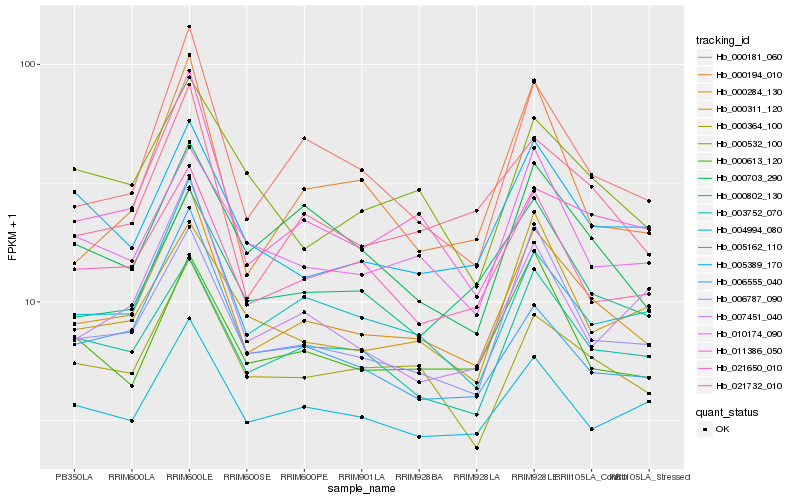
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_130 |
0.0 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 2 |
Hb_006787_090 |
0.0800345291 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 3 |
Hb_021732_010 |
0.0811396703 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 4 |
Hb_004994_080 |
0.0812984921 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000181_060 |
0.0827170271 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000364_100 |
0.0867493624 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 7 |
Hb_003752_070 |
0.0899032169 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 8 |
Hb_021650_010 |
0.0900790288 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 9 |
Hb_010174_090 |
0.1028049736 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005162_110 |
0.102870223 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 11 |
Hb_000613_120 |
0.105610007 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000802_130 |
0.1063262537 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 13 |
Hb_005389_170 |
0.1082251708 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 14 |
Hb_000703_290 |
0.1087307906 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 15 |
Hb_006555_040 |
0.1111666069 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 16 |
Hb_000194_010 |
0.111428533 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_011386_050 |
0.1115418695 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_007451_040 |
0.1129588892 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 19 |
Hb_000311_120 |
0.1132858835 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 20 |
Hb_000532_100 |
0.1136180504 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |