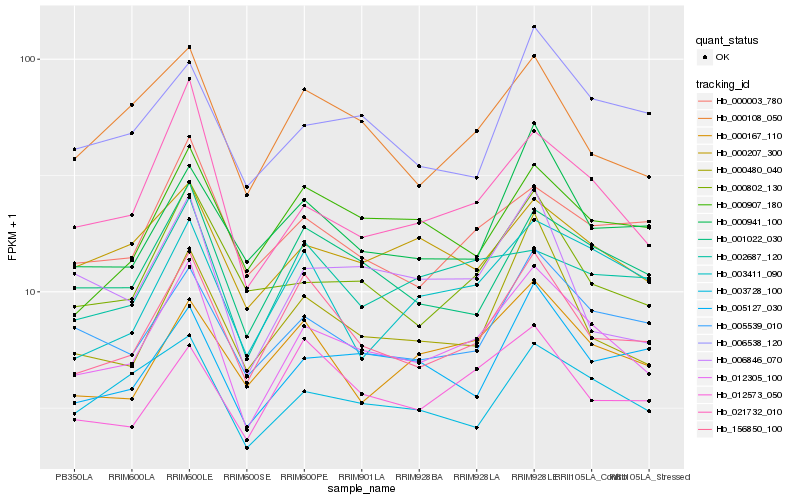
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012305_100 |
0.0 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_021732_010 |
0.0852015235 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 3 |
Hb_156850_100 |
0.0962540894 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 4 |
Hb_001022_030 |
0.0980308345 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 5 |
Hb_005539_010 |
0.1024550987 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 6 |
Hb_003728_100 |
0.1056738314 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000802_130 |
0.1062171115 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 8 |
Hb_000207_300 |
0.1069775997 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000108_050 |
0.1073920469 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 10 |
Hb_000480_040 |
0.1078143922 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 11 |
Hb_005127_030 |
0.1109978336 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_000003_780 |
0.1117702429 |
- |
- |
hexokinase [Manihot esculenta] |
| 13 |
Hb_000167_110 |
0.1151319218 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_006846_070 |
0.1152847736 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_002687_120 |
0.1155197587 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_012573_050 |
0.1173347281 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 17 |
Hb_000907_180 |
0.1177236078 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006538_120 |
0.1188399462 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003411_090 |
0.1188890676 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_000941_100 |
0.1189061302 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |