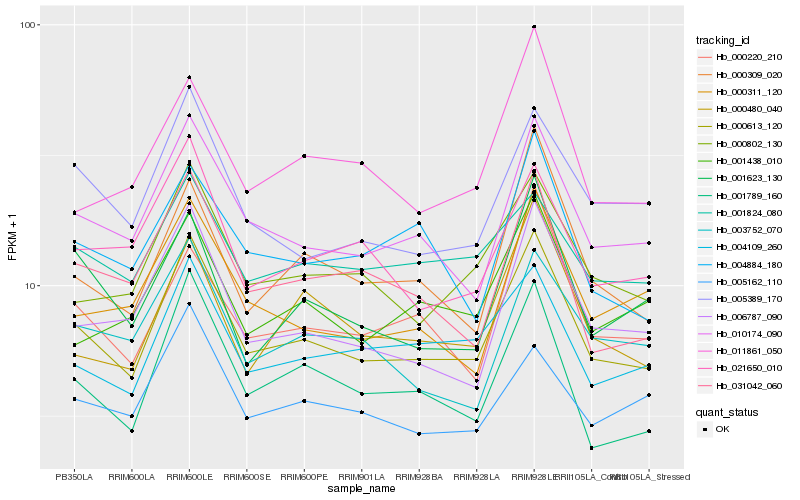
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_120 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_010174_090 |
0.0680666074 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000802_130 |
0.0801213665 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 4 |
Hb_001789_160 |
0.0818549707 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 5 |
Hb_006787_090 |
0.0835484337 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 6 |
Hb_004109_260 |
0.0848077988 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 7 |
Hb_031042_060 |
0.0889381258 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_005389_170 |
0.0902846408 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 9 |
Hb_021650_010 |
0.0911753287 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 10 |
Hb_000220_210 |
0.0915784833 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000309_020 |
0.0923133231 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 12 |
Hb_001438_010 |
0.0930041429 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000480_040 |
0.0943620609 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 14 |
Hb_001824_080 |
0.0947501805 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 15 |
Hb_003752_070 |
0.0954132333 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000311_120 |
0.0956606382 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 17 |
Hb_005162_110 |
0.096449991 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 18 |
Hb_001623_130 |
0.0971816825 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_011861_050 |
0.0980408377 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_004884_180 |
0.0999553741 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |