| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
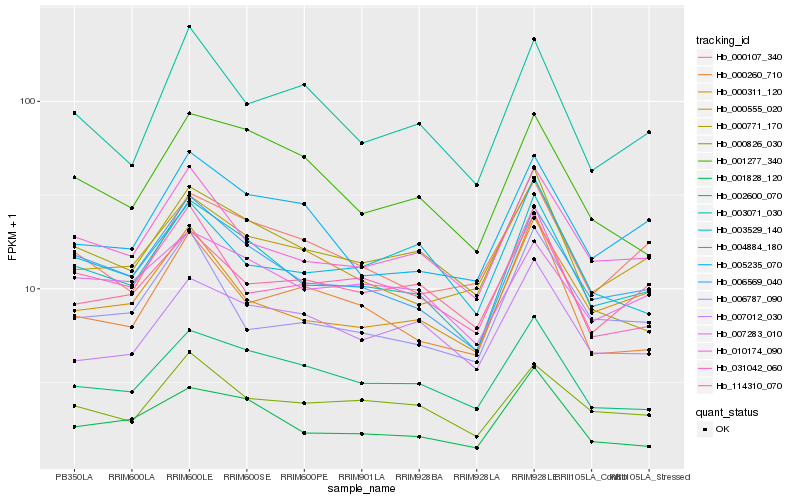
Hb_003529_140 |
0.0 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006569_040 |
0.0461419737 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 3 |
Hb_010174_090 |
0.0721261986 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001828_120 |
0.0782181008 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 5 |
Hb_002600_070 |
0.086693412 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 6 |
Hb_000555_020 |
0.0914228987 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000311_120 |
0.091450388 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 8 |
Hb_000826_030 |
0.0949266227 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 9 |
Hb_007283_010 |
0.0987921309 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 10 |
Hb_000771_170 |
0.0992426037 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_031042_060 |
0.099961595 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_005235_070 |
0.1009395544 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000107_340 |
0.1009938925 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 14 |
Hb_001277_340 |
0.1034542971 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 15 |
Hb_003071_030 |
0.1043326903 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 16 |
Hb_114310_070 |
0.1054659397 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 17 |
Hb_004884_180 |
0.1087494309 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000260_710 |
0.1096477467 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007012_030 |
0.1099575137 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_006787_090 |
0.1107683304 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |