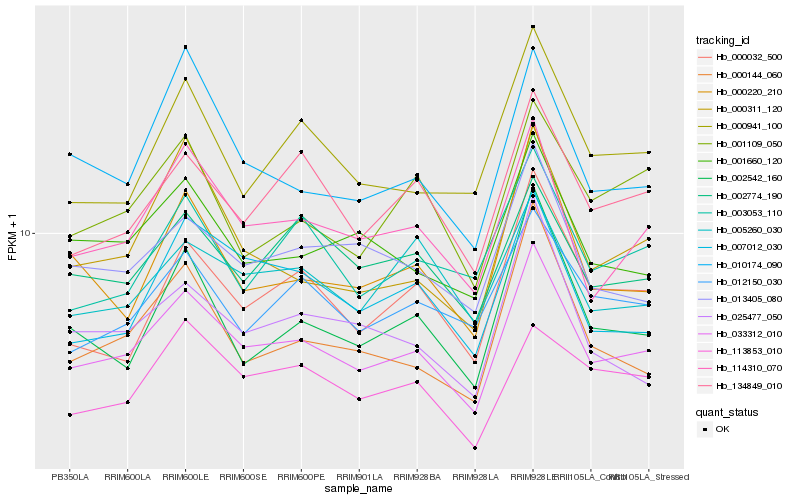
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_010 |
0.0 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_114310_070 |
0.0632069752 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 3 |
Hb_013405_080 |
0.0769996045 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 4 |
Hb_001660_120 |
0.0775583751 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_134849_010 |
0.0781518718 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 6 |
Hb_002774_190 |
0.0840810097 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 7 |
Hb_000032_500 |
0.0844539528 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 8 |
Hb_007012_030 |
0.0847179087 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_012150_030 |
0.084918259 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000941_100 |
0.0861500136 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 11 |
Hb_000144_060 |
0.0868481824 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000311_120 |
0.0886746999 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 13 |
Hb_002542_160 |
0.0889452486 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_025477_050 |
0.089886951 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_000220_210 |
0.0898959402 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005260_030 |
0.0904750257 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 17 |
Hb_010174_090 |
0.0907664381 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003053_110 |
0.0907803031 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_113853_010 |
0.0916162776 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 20 |
Hb_001109_050 |
0.0917538447 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |