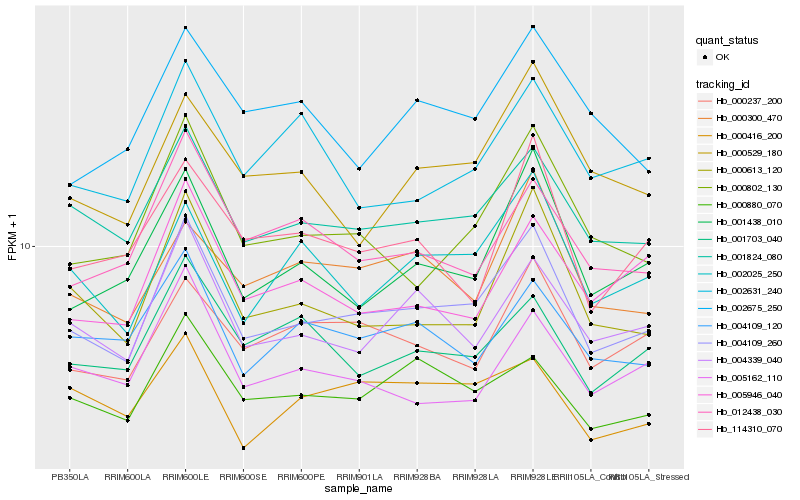
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_260 |
0.0 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 2 |
Hb_001824_080 |
0.0679689506 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 3 |
Hb_004339_040 |
0.0830922929 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |
| 4 |
Hb_000802_130 |
0.0846147091 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 5 |
Hb_000613_120 |
0.0848077988 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001438_010 |
0.086074083 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000880_070 |
0.0902797404 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 8 |
Hb_002025_250 |
0.0911589459 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 9 |
Hb_000237_200 |
0.0928128522 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 10 |
Hb_004109_120 |
0.0958106405 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012438_030 |
0.0983024699 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 12 |
Hb_000300_470 |
0.0988238757 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 13 |
Hb_000529_180 |
0.0989931507 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001703_040 |
0.0992866775 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005946_040 |
0.1006619395 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000416_200 |
0.1026927922 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 17 |
Hb_005162_110 |
0.1033869754 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 18 |
Hb_002675_250 |
0.1054484041 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 19 |
Hb_002631_240 |
0.1060674187 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 20 |
Hb_114310_070 |
0.1069872606 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |