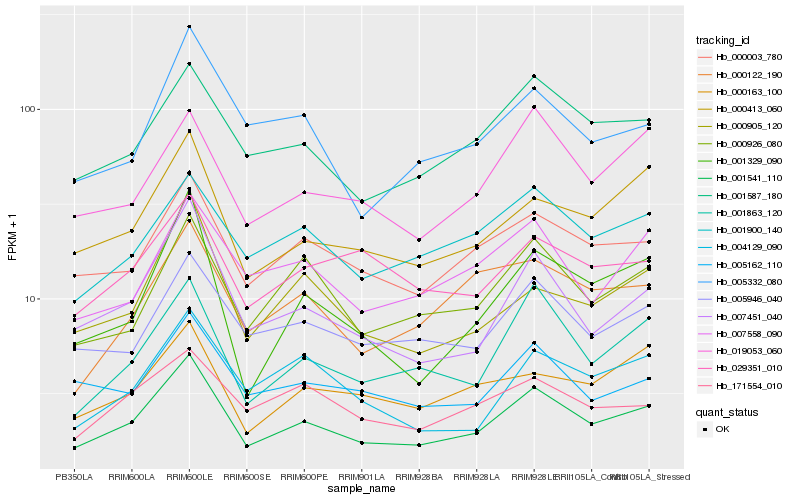
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_110 |
0.0 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_007451_040 |
0.0836206099 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 3 |
Hb_000003_780 |
0.0885279836 |
- |
- |
hexokinase [Manihot esculenta] |
| 4 |
Hb_001587_180 |
0.091052899 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 5 |
Hb_000413_060 |
0.0925731131 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 6 |
Hb_001863_120 |
0.0928709615 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000926_080 |
0.096725954 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 8 |
Hb_005946_040 |
0.0968289913 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001900_140 |
0.1027830404 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000905_120 |
0.1045818008 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 11 |
Hb_019053_060 |
0.1060931884 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 12 |
Hb_001329_090 |
0.10774377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007558_090 |
0.1078003005 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_005162_110 |
0.1084065317 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 15 |
Hb_005332_080 |
0.1088841953 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000163_100 |
0.1116896549 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 17 |
Hb_004129_090 |
0.1119622735 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 18 |
Hb_171554_010 |
0.1141458467 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 19 |
Hb_029351_010 |
0.1145039996 |
- |
- |
PREDICTED: HD domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000122_190 |
0.1146676297 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |