| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
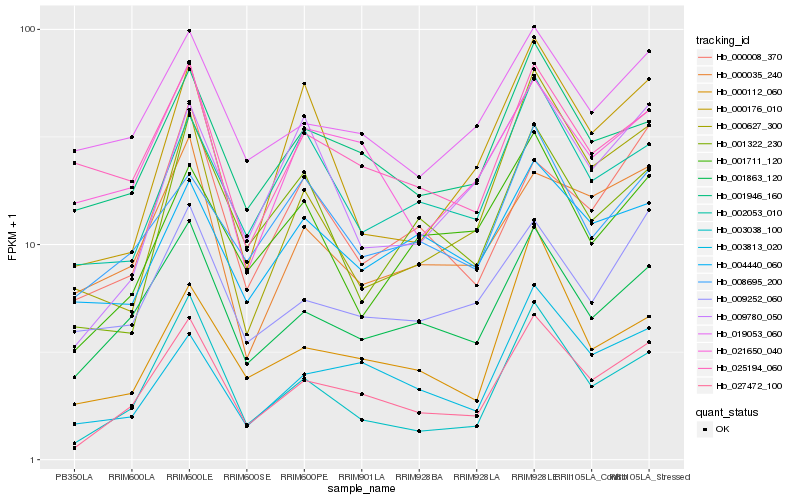
Hb_027472_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 2 |
Hb_001863_120 |
0.100433457 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003038_100 |
0.1115535704 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001946_160 |
0.1226056074 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 5 |
Hb_003813_020 |
0.1268198667 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 6 |
Hb_021650_040 |
0.1374533556 |
- |
- |
EG2771 [Manihot esculenta] |
| 7 |
Hb_009252_060 |
0.1384617143 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 8 |
Hb_000035_240 |
0.139990086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_019053_060 |
0.1409887449 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 10 |
Hb_009780_050 |
0.1446557268 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000112_060 |
0.145835866 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002053_010 |
0.1460233301 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000008_370 |
0.1473252697 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 14 |
Hb_000627_300 |
0.1488795145 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 15 |
Hb_008695_200 |
0.1494988682 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 16 |
Hb_000176_010 |
0.1496450051 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 17 |
Hb_025194_060 |
0.1508942795 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_001711_120 |
0.1524983428 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004440_060 |
0.1531461986 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 20 |
Hb_001322_230 |
0.1545402827 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |