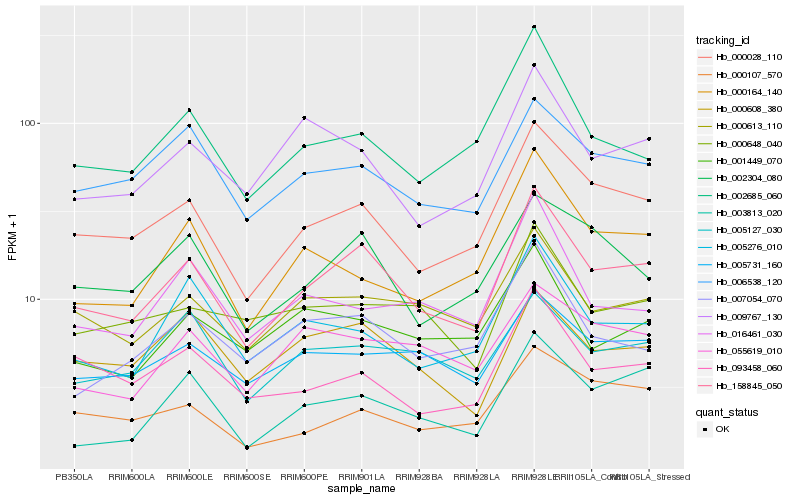
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158845_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 2 |
Hb_000028_110 |
0.0842542324 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 3 |
Hb_000613_110 |
0.1118470362 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 4 |
Hb_002304_080 |
0.116099532 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |
| 5 |
Hb_000608_380 |
0.120920669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005731_160 |
0.1210118283 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_006538_120 |
0.1210487032 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003813_020 |
0.1222814833 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002685_060 |
0.1227247375 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 10 |
Hb_005276_010 |
0.1235824106 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 11 |
Hb_007054_070 |
0.1241539556 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 12 |
Hb_009767_130 |
0.1263450605 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 13 |
Hb_093458_060 |
0.1273149756 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 14 |
Hb_005127_030 |
0.1289469134 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 15 |
Hb_000648_040 |
0.1292271509 |
- |
- |
unknown [Populus trichocarpa] |
| 16 |
Hb_000107_570 |
0.1297482011 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001449_070 |
0.1301194617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_055619_010 |
0.1308267255 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000164_140 |
0.1321622641 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_016461_030 |
0.1341302322 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |