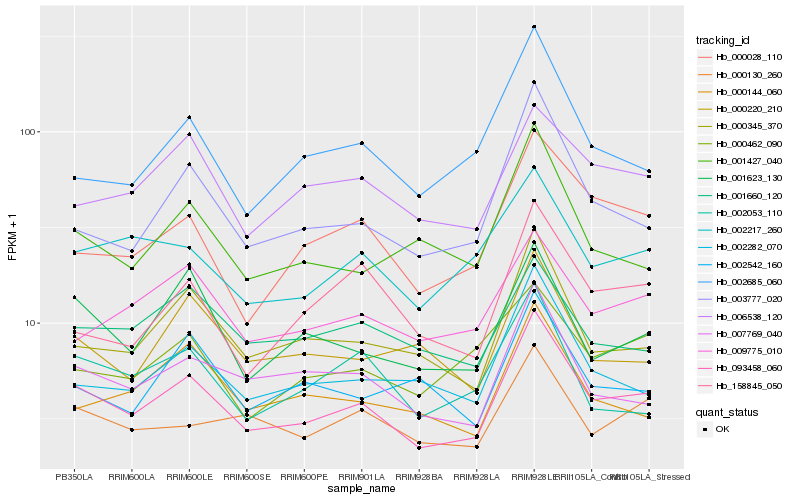
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093458_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 2 |
Hb_000345_370 |
0.1028101522 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 3 |
Hb_007769_040 |
0.1052743035 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_003777_020 |
0.1056082301 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 5 |
Hb_000028_110 |
0.1100173171 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 6 |
Hb_002282_070 |
0.1100788394 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 7 |
Hb_001623_130 |
0.1129876034 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002217_260 |
0.1144249573 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 9 |
Hb_000130_260 |
0.1179815143 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006538_120 |
0.1194444124 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001427_040 |
0.1195932434 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 12 |
Hb_000220_210 |
0.1212716663 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009775_010 |
0.1218144724 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 14 |
Hb_002685_060 |
0.1226463177 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 15 |
Hb_000144_060 |
0.123867725 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_002053_110 |
0.1239742798 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001660_120 |
0.1254132398 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_158845_050 |
0.1273149756 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 19 |
Hb_002542_160 |
0.1278950311 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000462_090 |
0.1282790853 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |