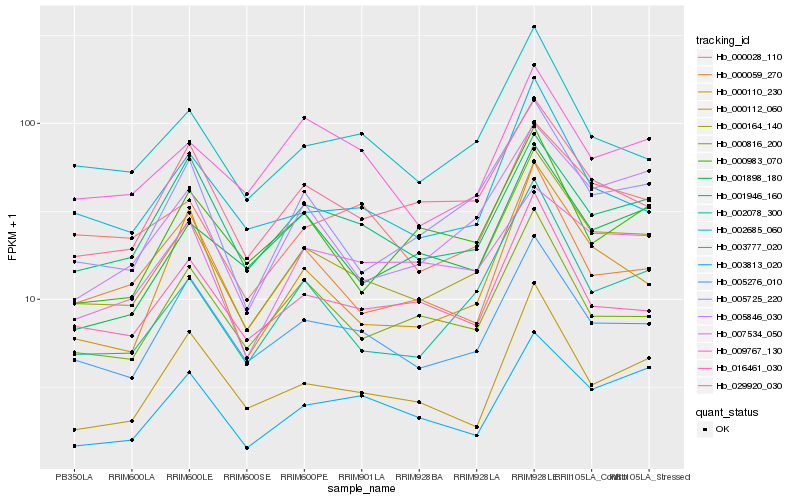
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000164_140 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_005725_220 |
0.0826089002 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 3 |
Hb_005276_010 |
0.0867504567 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 4 |
Hb_002078_300 |
0.0933352818 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005846_030 |
0.0955897538 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 6 |
Hb_029920_030 |
0.0957343945 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000028_110 |
0.1055194124 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 8 |
Hb_002685_060 |
0.1056434395 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 9 |
Hb_001946_160 |
0.1072737017 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 10 |
Hb_000110_230 |
0.1089983503 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000816_200 |
0.1092698367 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 12 |
Hb_003777_020 |
0.1092759065 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 13 |
Hb_016461_030 |
0.1115926563 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000059_270 |
0.1133092241 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 15 |
Hb_001898_180 |
0.1162452562 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000112_060 |
0.1187124685 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003813_020 |
0.122024184 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000983_070 |
0.1223548664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007534_050 |
0.1230471766 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_009767_130 |
0.1238767526 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |