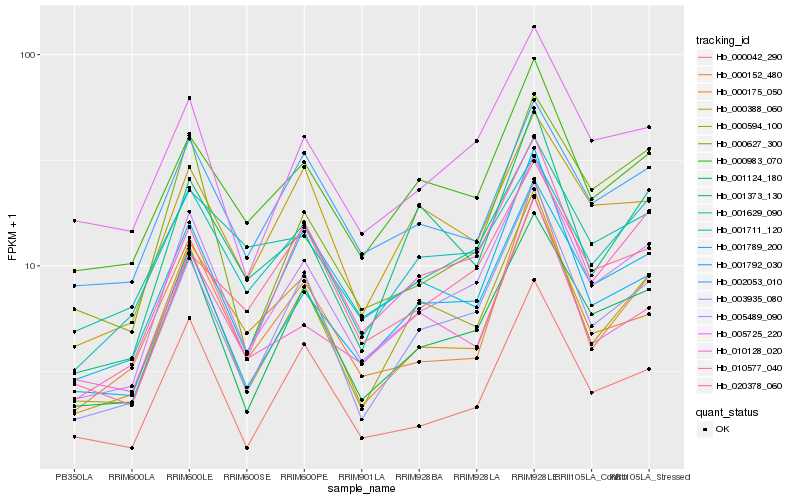
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005489_090 |
0.0 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001124_180 |
0.0634350216 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 3 |
Hb_003935_080 |
0.0694102082 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000594_100 |
0.0835081643 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_010577_040 |
0.0878565337 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 6 |
Hb_005725_220 |
0.0956510263 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 7 |
Hb_001792_030 |
0.0994458925 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 8 |
Hb_000983_070 |
0.1003682315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000042_290 |
0.1028295284 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000152_480 |
0.1035242192 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000175_050 |
0.1086761452 |
- |
- |
- |
| 12 |
Hb_001711_120 |
0.1087509428 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000627_300 |
0.112067906 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 14 |
Hb_020378_060 |
0.1161149005 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 15 |
Hb_000388_060 |
0.1169215786 |
- |
- |
fructokinase [Manihot esculenta] |
| 16 |
Hb_010128_020 |
0.1172949035 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 17 |
Hb_001789_200 |
0.1179577271 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001373_130 |
0.1192284603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002053_010 |
0.119273283 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001629_090 |
0.1223883444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |