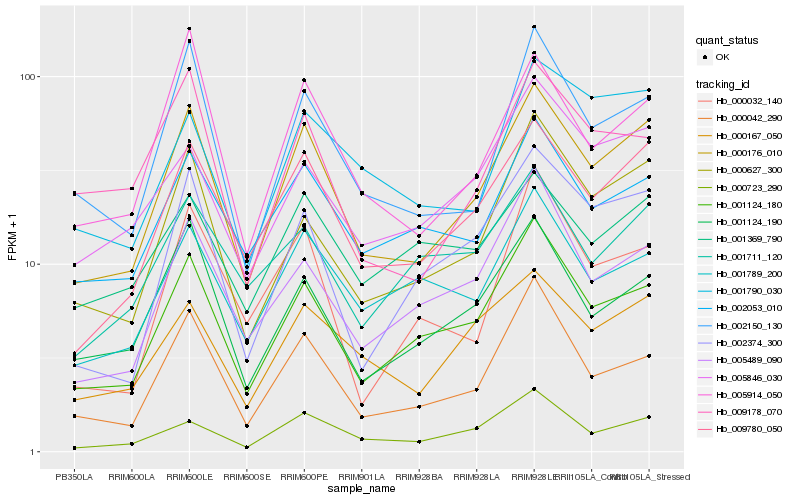
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000176_010 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 2 |
Hb_009780_050 |
0.074371258 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001124_180 |
0.0944618098 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 4 |
Hb_000627_300 |
0.1087219877 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 5 |
Hb_000042_290 |
0.1096475998 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002150_130 |
0.1108814396 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000167_050 |
0.1138845205 |
- |
- |
PREDICTED: uncharacterized protein LOC105641106 [Jatropha curcas] |
| 8 |
Hb_005914_050 |
0.1146087037 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001124_190 |
0.1163794851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002374_300 |
0.1167569404 |
- |
- |
hypothetical protein JCGZ_12140 [Jatropha curcas] |
| 11 |
Hb_000723_290 |
0.1172237377 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_002053_010 |
0.1225193178 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_005489_090 |
0.1268528956 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_005846_030 |
0.1323590391 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001789_200 |
0.1349738015 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 16 |
Hb_009178_070 |
0.1357536983 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001790_030 |
0.1413977309 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001711_120 |
0.1414713954 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000032_140 |
0.1442768135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001369_790 |
0.1458403408 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |