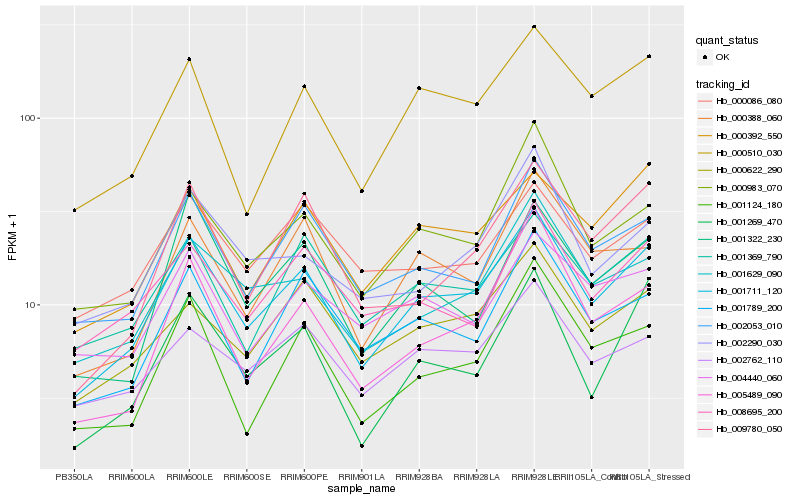
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001711_120 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000510_030 |
0.0905123744 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 3 |
Hb_002053_010 |
0.0917626049 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_001629_090 |
0.0927980046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000983_070 |
0.0952202502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000392_550 |
0.0974304048 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_001369_790 |
0.0984762706 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000622_290 |
0.0988676571 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 9 |
Hb_001789_200 |
0.1017513891 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002762_110 |
0.1036113676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000388_060 |
0.1041776465 |
- |
- |
fructokinase [Manihot esculenta] |
| 12 |
Hb_009780_050 |
0.1054841096 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_008695_200 |
0.107764321 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 14 |
Hb_005489_090 |
0.1087509428 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001269_470 |
0.1095201939 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 16 |
Hb_001124_180 |
0.1098642587 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 17 |
Hb_001322_230 |
0.1110903819 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 18 |
Hb_004440_060 |
0.1132391153 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 19 |
Hb_002290_030 |
0.1138898083 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000086_080 |
0.1145945769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |