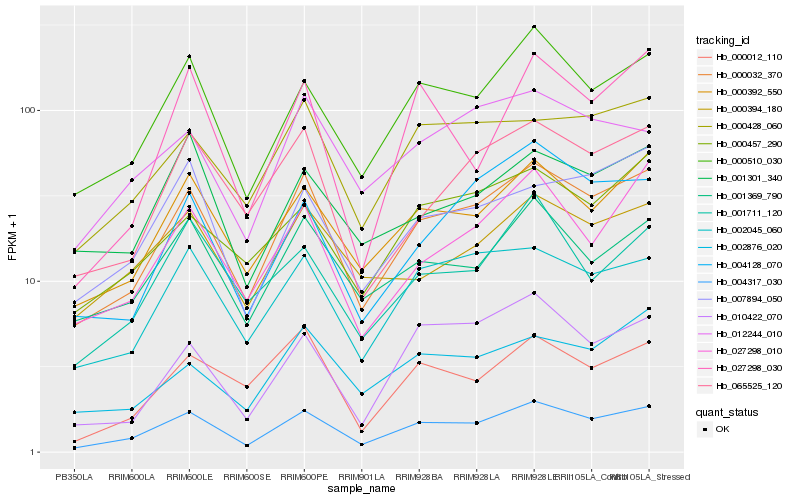
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004317_030 |
0.0 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 2 |
Hb_000032_370 |
0.0490067593 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 3 |
Hb_000510_030 |
0.0766768749 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 4 |
Hb_000392_550 |
0.0905120622 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_002045_060 |
0.1052492287 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001369_790 |
0.1083749514 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007894_050 |
0.1158883721 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000428_060 |
0.1159802598 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_027298_010 |
0.1214897265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_065525_120 |
0.1230835356 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 11 |
Hb_010422_070 |
0.1238396858 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |
| 12 |
Hb_001711_120 |
0.1238843852 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000457_290 |
0.1255755503 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 14 |
Hb_000394_180 |
0.1259287401 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 15 |
Hb_001301_340 |
0.1278592323 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 16 |
Hb_012244_010 |
0.127958908 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 17 |
Hb_000012_110 |
0.1280150947 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 18 |
Hb_027298_030 |
0.130974789 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 19 |
Hb_002876_020 |
0.1311902049 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004128_070 |
0.1318205129 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |