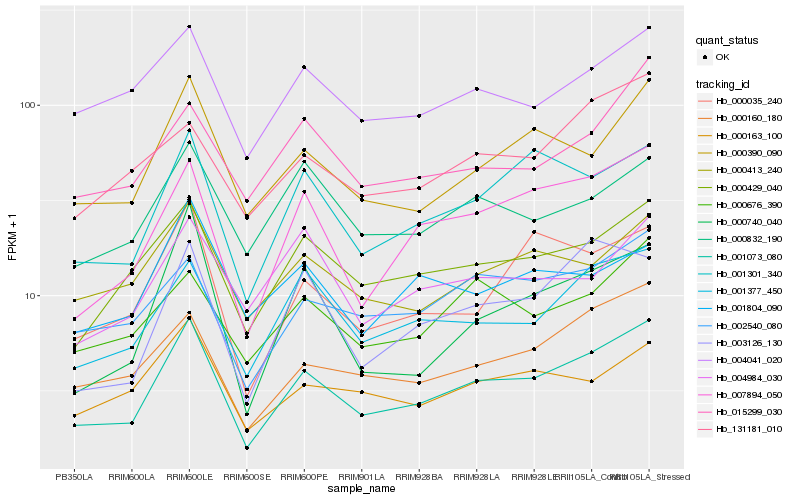
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001073_080 |
0.0 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007894_050 |
0.0907946075 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000390_090 |
0.1007081763 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 4 |
Hb_001301_340 |
0.1029833296 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 5 |
Hb_001377_450 |
0.1036724712 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000160_180 |
0.1044326291 |
- |
- |
methionyl-tRNA formyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000429_040 |
0.1048987995 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 8 |
Hb_003126_130 |
0.1125537782 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 9 |
Hb_002540_080 |
0.113456294 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 10 |
Hb_000035_240 |
0.1152353858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000740_040 |
0.1189164669 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 12 |
Hb_000413_240 |
0.1222434741 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000676_390 |
0.1231967553 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000163_100 |
0.1243058046 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 15 |
Hb_004041_020 |
0.1249491962 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 16 |
Hb_015299_030 |
0.1262003903 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 17 |
Hb_000832_190 |
0.128742918 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 18 |
Hb_131181_010 |
0.1304855941 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 19 |
Hb_004984_030 |
0.1344422158 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_001804_090 |
0.1388131996 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |