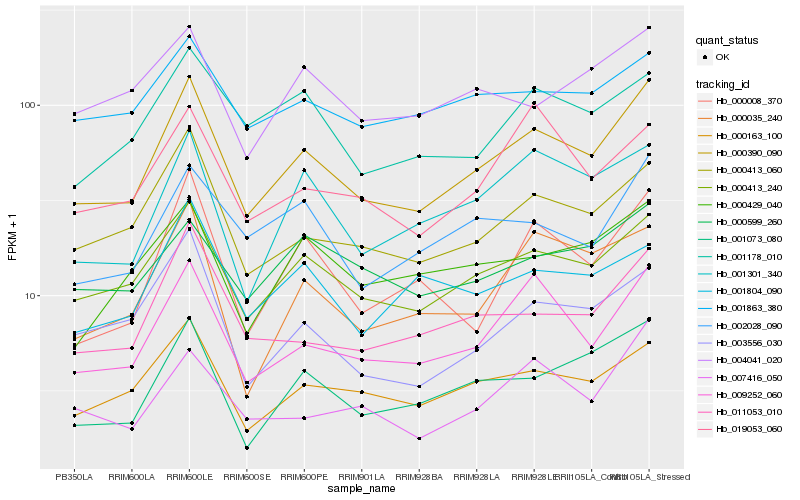
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 2 |
Hb_000413_240 |
0.0711000455 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_009252_060 |
0.0860828926 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 4 |
Hb_000413_060 |
0.0877393465 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 5 |
Hb_000163_100 |
0.0924932087 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 6 |
Hb_001073_080 |
0.1007081763 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011053_010 |
0.1010717052 |
- |
- |
PREDICTED: uncharacterized protein LOC105631146 [Jatropha curcas] |
| 8 |
Hb_002028_090 |
0.107400871 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 9 |
Hb_001301_340 |
0.108160372 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 10 |
Hb_003556_030 |
0.1115623623 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 11 |
Hb_000035_240 |
0.1120003063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007416_050 |
0.1146639235 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 13 |
Hb_000008_370 |
0.1147642333 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 14 |
Hb_001178_010 |
0.1162050232 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 15 |
Hb_000429_040 |
0.1177099204 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 16 |
Hb_019053_060 |
0.1208695353 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 17 |
Hb_001863_380 |
0.1213344352 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 18 |
Hb_000599_260 |
0.1216447642 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 19 |
Hb_001804_090 |
0.1238701967 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |
| 20 |
Hb_004041_020 |
0.1245021038 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |