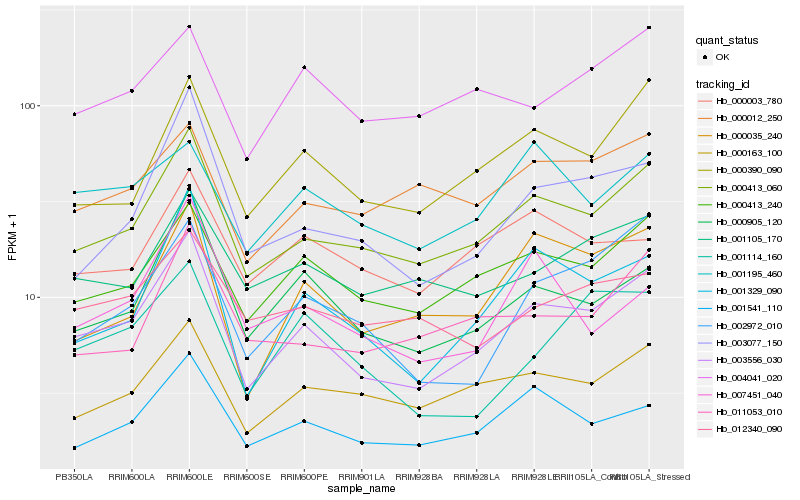
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003556_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 2 |
Hb_000413_060 |
0.0735538249 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 3 |
Hb_001329_090 |
0.1052154614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000905_120 |
0.1062364502 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 5 |
Hb_000413_240 |
0.1089281231 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000390_090 |
0.1115623623 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 7 |
Hb_003077_150 |
0.1125685502 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 8 |
Hb_001541_110 |
0.1149310462 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_000163_100 |
0.1161609947 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 10 |
Hb_000035_240 |
0.1232623604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002972_010 |
0.1293159214 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 12 |
Hb_001105_170 |
0.132427486 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 13 |
Hb_011053_010 |
0.1332719152 |
- |
- |
PREDICTED: uncharacterized protein LOC105631146 [Jatropha curcas] |
| 14 |
Hb_001114_160 |
0.1408332078 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 15 |
Hb_012340_090 |
0.1410512751 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 16 |
Hb_000012_250 |
0.1416169142 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 17 |
Hb_004041_020 |
0.1424387888 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 18 |
Hb_007451_040 |
0.1432508458 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 19 |
Hb_001195_460 |
0.1457726773 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000003_780 |
0.146056627 |
- |
- |
hexokinase [Manihot esculenta] |