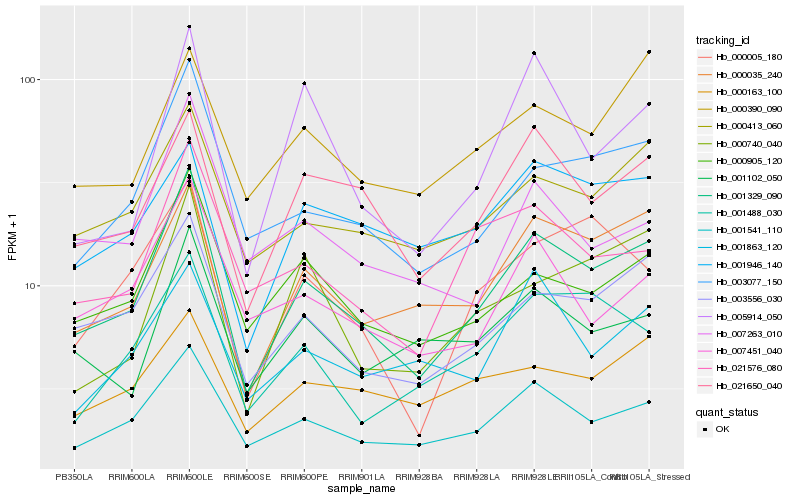
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003556_030 |
0.1052154614 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 3 |
Hb_003077_150 |
0.1061215972 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 4 |
Hb_001541_110 |
0.10774377 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_000035_240 |
0.1109407106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000905_120 |
0.1174912249 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 7 |
Hb_000413_060 |
0.1220985566 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 8 |
Hb_021650_040 |
0.124182021 |
- |
- |
EG2771 [Manihot esculenta] |
| 9 |
Hb_021576_080 |
0.1333911932 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 10 |
Hb_007451_040 |
0.1352898133 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 11 |
Hb_000740_040 |
0.1382477757 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 12 |
Hb_001102_050 |
0.1382697647 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 13 |
Hb_000005_180 |
0.1403852695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001488_030 |
0.1423478824 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 15 |
Hb_005914_050 |
0.1438837657 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000390_090 |
0.1452676193 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 17 |
Hb_007263_010 |
0.1463707268 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_000163_100 |
0.1473909369 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 19 |
Hb_001946_140 |
0.1474882435 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001863_120 |
0.1513445386 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |