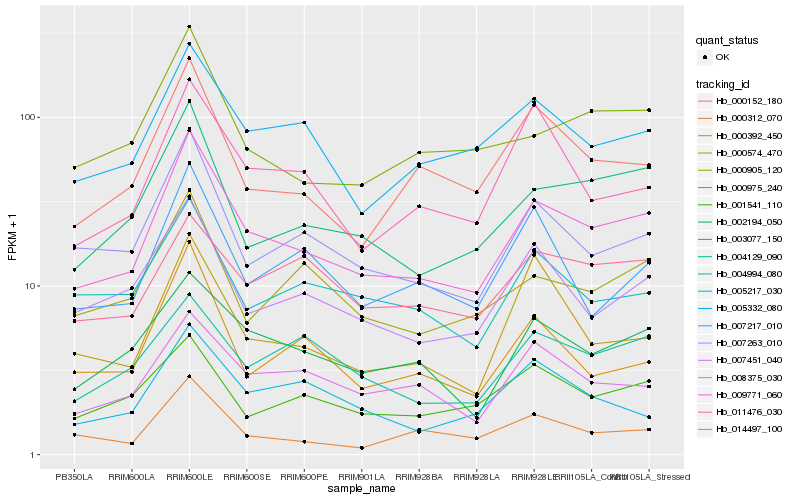
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009771_060 |
0.0 |
- |
- |
PREDICTED: 30S ribosomal protein S31, chloroplastic [Populus euphratica] |
| 2 |
Hb_003077_150 |
0.1051656318 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 3 |
Hb_007263_010 |
0.1079666546 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 4 |
Hb_002194_050 |
0.1115974527 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005332_080 |
0.1137853747 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_007451_040 |
0.1161879644 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 7 |
Hb_000152_180 |
0.1206534273 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 8 |
Hb_008375_030 |
0.1225957949 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 9 |
Hb_000975_240 |
0.1233682757 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 10 |
Hb_000905_120 |
0.1266995872 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 11 |
Hb_001541_110 |
0.129413072 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_004994_080 |
0.1323820121 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_004129_090 |
0.1332842206 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 14 |
Hb_011476_030 |
0.1367536862 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 15 |
Hb_007217_010 |
0.1375216606 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000312_070 |
0.1385660433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_014497_100 |
0.1391021552 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000392_450 |
0.1415242233 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000574_470 |
0.1455255179 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_005217_030 |
0.1463317726 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |