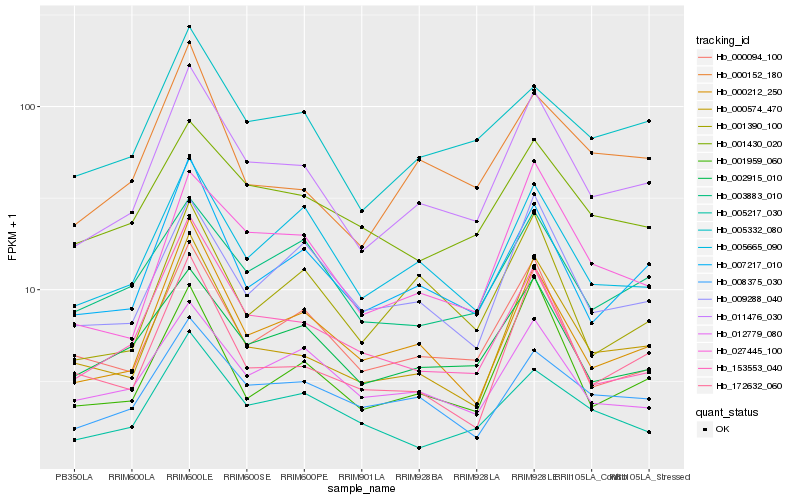
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011476_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 2 |
Hb_007217_010 |
0.0930347344 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000574_470 |
0.0950539269 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000212_250 |
0.096716795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_027445_100 |
0.1012601051 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 6 |
Hb_002915_010 |
0.1061172962 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 7 |
Hb_005665_090 |
0.1065250775 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 8 |
Hb_008375_030 |
0.1070196169 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 9 |
Hb_000094_100 |
0.1084151148 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 10 |
Hb_000152_180 |
0.1125584573 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 11 |
Hb_005332_080 |
0.1132042749 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_009288_040 |
0.1151406187 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 13 |
Hb_153553_040 |
0.1158336261 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 14 |
Hb_001430_020 |
0.116255102 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_012779_080 |
0.116776862 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 16 |
Hb_172632_060 |
0.1178865442 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001959_060 |
0.1179683812 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003883_010 |
0.1185837964 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 19 |
Hb_005217_030 |
0.1199246435 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 20 |
Hb_001390_100 |
0.1221716453 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |