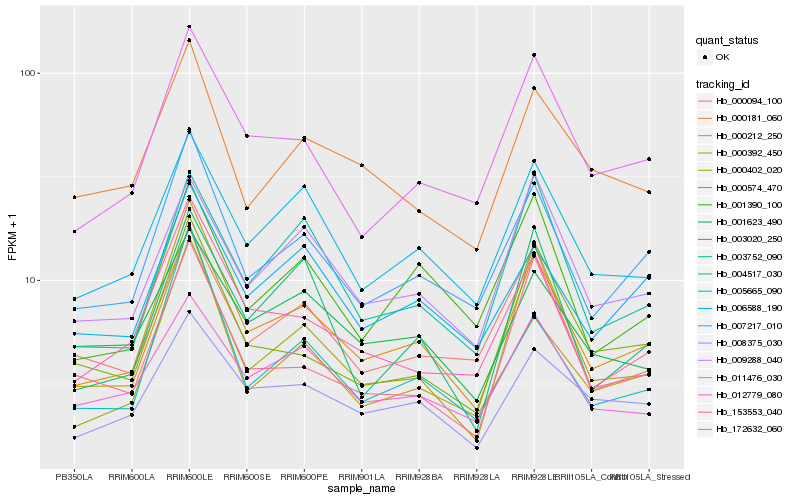
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007217_010 |
0.0693062625 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004517_030 |
0.0876901461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_153553_040 |
0.0961109823 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 5 |
Hb_011476_030 |
0.096716795 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 6 |
Hb_000392_450 |
0.1007628533 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005665_090 |
0.101564343 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 8 |
Hb_008375_030 |
0.1051569773 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 9 |
Hb_000574_470 |
0.1083189154 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_000402_020 |
0.1091075282 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000181_060 |
0.1134790468 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000094_100 |
0.1143025827 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 13 |
Hb_012779_080 |
0.1170691509 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 14 |
Hb_006588_190 |
0.1175874592 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 15 |
Hb_172632_060 |
0.1177831117 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003752_090 |
0.118642937 |
- |
- |
chitinase, putative [Ricinus communis] |
| 17 |
Hb_001390_100 |
0.120951911 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001623_490 |
0.1210190945 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009288_040 |
0.1211222506 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 20 |
Hb_003020_250 |
0.1214346095 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |