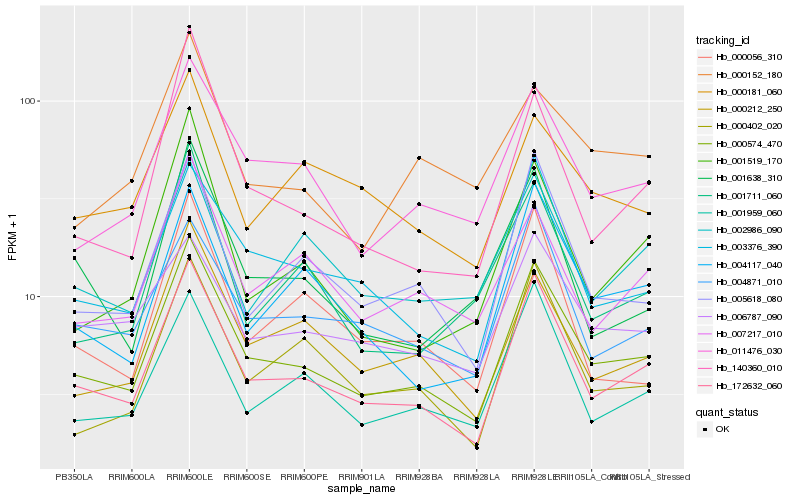
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_470 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_172632_060 |
0.0461448435 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011476_030 |
0.0950539269 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 4 |
Hb_003376_390 |
0.1042133801 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005618_080 |
0.104937135 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000212_250 |
0.1083189154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001959_060 |
0.1107571956 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004871_010 |
0.1141714908 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 9 |
Hb_140360_010 |
0.1190801885 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_004117_040 |
0.1192281672 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 11 |
Hb_007217_010 |
0.1196286058 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000402_020 |
0.1212296679 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_006787_090 |
0.1233784374 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 14 |
Hb_001638_310 |
0.1268188682 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 15 |
Hb_001519_170 |
0.1274898248 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001711_060 |
0.1288013723 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 17 |
Hb_000152_180 |
0.1315995374 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 18 |
Hb_000181_060 |
0.1317492172 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002986_090 |
0.1332133053 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_000056_310 |
0.1342245976 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |