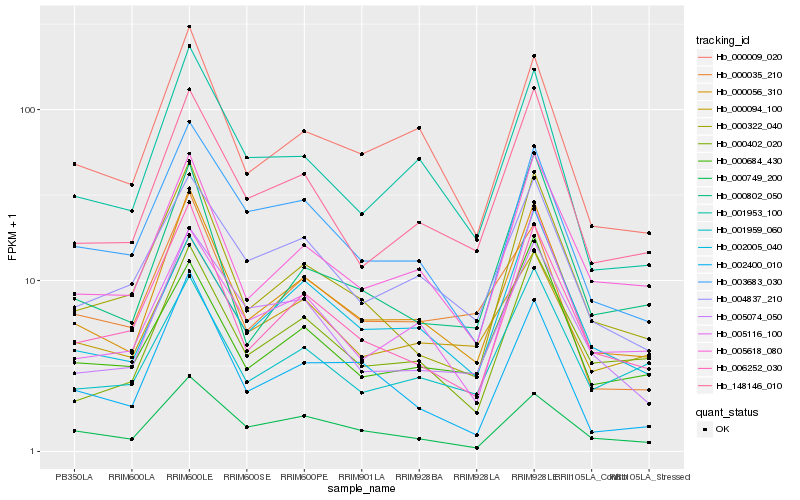
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 2 |
Hb_148146_010 |
0.0817412405 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 3 |
Hb_006252_030 |
0.0846416611 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 4 |
Hb_005618_080 |
0.0863950421 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000009_020 |
0.0870298468 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001953_100 |
0.0927123398 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000749_200 |
0.0987094203 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000322_040 |
0.1014101315 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000402_020 |
0.1019973807 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_002005_040 |
0.1077814676 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_003683_030 |
0.1081183302 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 12 |
Hb_004837_210 |
0.1086997188 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 13 |
Hb_000094_100 |
0.1103666637 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 14 |
Hb_000802_050 |
0.1110919281 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 15 |
Hb_001959_060 |
0.1127308967 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000684_430 |
0.113804645 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005116_100 |
0.1149417588 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 18 |
Hb_000035_210 |
0.1158558783 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 19 |
Hb_005074_050 |
0.1164341381 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 20 |
Hb_002400_010 |
0.116607585 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |