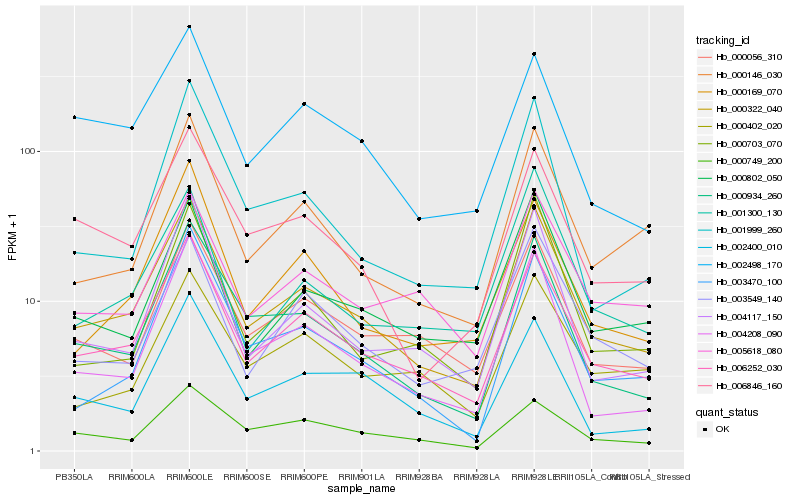
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000322_040 |
0.0 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006252_030 |
0.0483518597 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 3 |
Hb_000934_260 |
0.0937146572 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000056_310 |
0.1014101315 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 5 |
Hb_004208_090 |
0.1016412361 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 6 |
Hb_000146_030 |
0.1072835185 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000802_050 |
0.1074912241 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 8 |
Hb_003549_140 |
0.1105871503 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_004117_150 |
0.1124934854 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 10 |
Hb_005618_080 |
0.1143113159 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006846_160 |
0.1158912312 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 12 |
Hb_000169_070 |
0.1166564203 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 13 |
Hb_001300_130 |
0.1217514923 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 14 |
Hb_003470_100 |
0.1219202718 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_002498_170 |
0.1222720743 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000703_070 |
0.1226355829 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002400_010 |
0.1231198487 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 18 |
Hb_001999_260 |
0.1236225964 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000749_200 |
0.1237889905 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000402_020 |
0.125281009 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |