| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
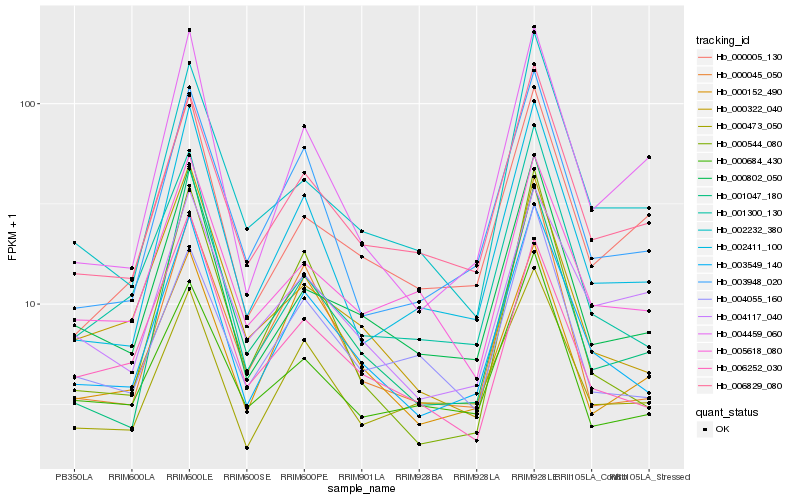
Hb_003549_140 |
0.0 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000802_050 |
0.0899882572 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 3 |
Hb_006829_080 |
0.0904051937 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 4 |
Hb_003948_020 |
0.0990540084 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 5 |
Hb_002411_100 |
0.100185796 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 6 |
Hb_001300_130 |
0.1059706597 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 7 |
Hb_000045_050 |
0.1071323379 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000473_050 |
0.1082382867 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 9 |
Hb_000322_040 |
0.1105871503 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006252_030 |
0.115965923 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 11 |
Hb_000684_430 |
0.116636016 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002232_380 |
0.1167897082 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_004055_160 |
0.1171996066 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005618_080 |
0.1186319075 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004459_060 |
0.119820234 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001047_180 |
0.1200532938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000544_080 |
0.1203817478 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000005_130 |
0.1215770277 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 19 |
Hb_000152_490 |
0.1223648604 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 20 |
Hb_004117_040 |
0.1224647789 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |